Zhiping Wang
USER: A Unified Information Search and Recommendation Model based on Integrated Behavior Sequence
Sep 30, 2021

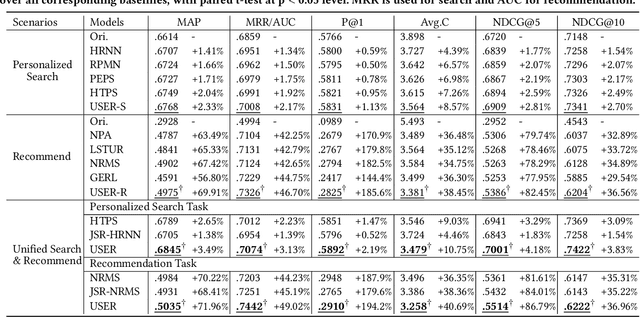
Abstract:Search and recommendation are the two most common approaches used by people to obtain information. They share the same goal -- satisfying the user's information need at the right time. There are already a lot of Internet platforms and Apps providing both search and recommendation services, showing us the demand and opportunity to simultaneously handle both tasks. However, most platforms consider these two tasks independently -- they tend to train separate search model and recommendation model, without exploiting the relatedness and dependency between them. In this paper, we argue that jointly modeling these two tasks will benefit both of them and finally improve overall user satisfaction. We investigate the interactions between these two tasks in the specific information content service domain. We propose first integrating the user's behaviors in search and recommendation into a heterogeneous behavior sequence, then utilizing a joint model for handling both tasks based on the unified sequence. More specifically, we design the Unified Information Search and Recommendation model (USER), which mines user interests from the integrated sequence and accomplish the two tasks in a unified way.
Uncovering protein interaction in abstracts and text using a novel linear model and word proximity networks
Dec 04, 2008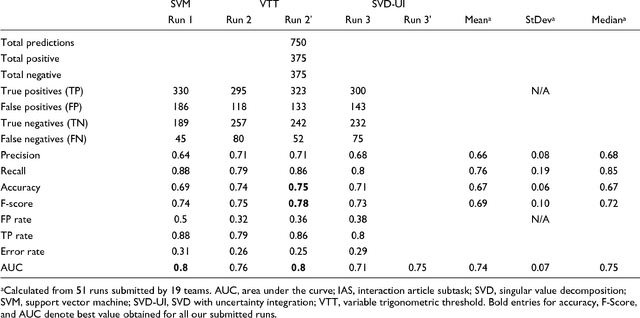
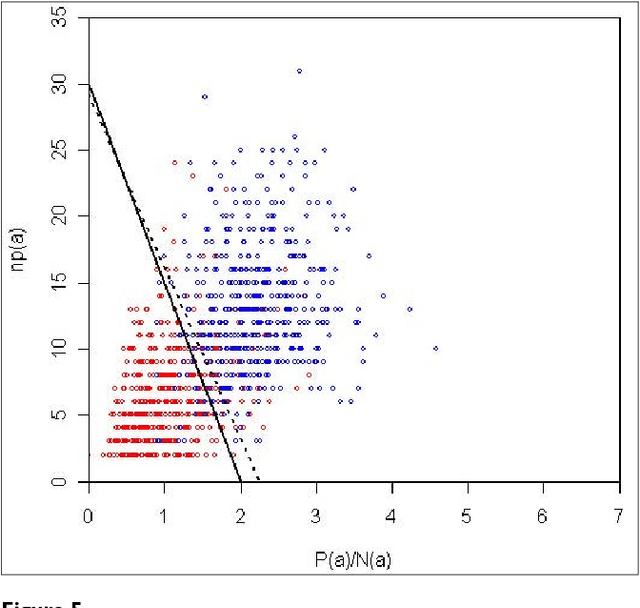
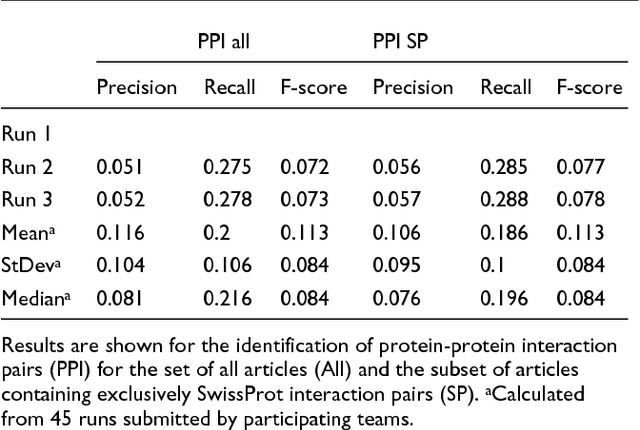
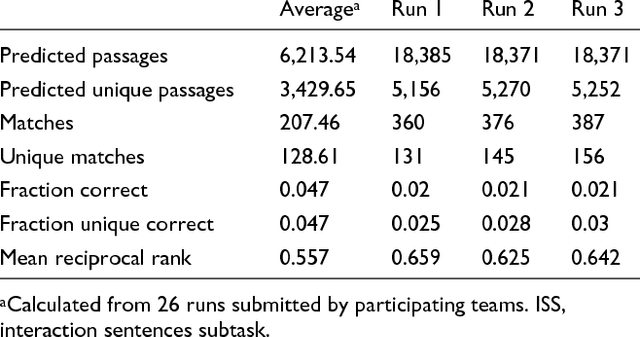
Abstract:We participated in three of the protein-protein interaction subtasks of the Second BioCreative Challenge: classification of abstracts relevant for protein-protein interaction (IAS), discovery of protein pairs (IPS) and text passages characterizing protein interaction (ISS) in full text documents. We approached the abstract classification task with a novel, lightweight linear model inspired by spam-detection techniques, as well as an uncertainty-based integration scheme. We also used a Support Vector Machine and the Singular Value Decomposition on the same features for comparison purposes. Our approach to the full text subtasks (protein pair and passage identification) includes a feature expansion method based on word-proximity networks. Our approach to the abstract classification task (IAS) was among the top submissions for this task in terms of the measures of performance used in the challenge evaluation (accuracy, F-score and AUC). We also report on a web-tool we produced using our approach: the Protein Interaction Abstract Relevance Evaluator (PIARE). Our approach to the full text tasks resulted in one of the highest recall rates as well as mean reciprocal rank of correct passages. Our approach to abstract classification shows that a simple linear model, using relatively few features, is capable of generalizing and uncovering the conceptual nature of protein-protein interaction from the bibliome. Since the novel approach is based on a very lightweight linear model, it can be easily ported and applied to similar problems. In full text problems, the expansion of word features with word-proximity networks is shown to be useful, though the need for some improvements is discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge