Zhiguo Huang
Step-GUI Technical Report
Dec 19, 2025



Abstract:Recent advances in multimodal large language models unlock unprecedented opportunities for GUI automation. However, a fundamental challenge remains: how to efficiently acquire high-quality training data while maintaining annotation reliability? We introduce a self-evolving training pipeline powered by the Calibrated Step Reward System, which converts model-generated trajectories into reliable training signals through trajectory-level calibration, achieving >90% annotation accuracy with 10-100x lower cost. Leveraging this pipeline, we introduce Step-GUI, a family of models (4B/8B) that achieves state-of-the-art GUI performance (8B: 80.2% AndroidWorld, 48.5% OSWorld, 62.6% ScreenShot-Pro) while maintaining robust general capabilities. As GUI agent capabilities improve, practical deployment demands standardized interfaces across heterogeneous devices while protecting user privacy. To this end, we propose GUI-MCP, the first Model Context Protocol for GUI automation with hierarchical architecture that combines low-level atomic operations and high-level task delegation to local specialist models, enabling high-privacy execution where sensitive data stays on-device. Finally, to assess whether agents can handle authentic everyday usage, we introduce AndroidDaily, a benchmark grounded in real-world mobile usage patterns with 3146 static actions and 235 end-to-end tasks across high-frequency daily scenarios (8B: static 89.91%, end-to-end 52.50%). Our work advances the development of practical GUI agents and demonstrates strong potential for real-world deployment in everyday digital interactions.
An artificially intelligent magnetic resonance spectroscopy quantification method: Comparison between QNet and LCModel on the cloud computing platform CloudBrain-MRS
Mar 06, 2025



Abstract:Objctives: This work aimed to statistically compare the metabolite quantification of human brain magnetic resonance spectroscopy (MRS) between the deep learning method QNet and the classical method LCModel through an easy-to-use intelligent cloud computing platform CloudBrain-MRS. Materials and Methods: In this retrospective study, two 3 T MRI scanners Philips Ingenia and Achieva collected 61 and 46 in vivo 1H magnetic resonance (MR) spectra of healthy participants, respectively, from the brain region of pregenual anterior cingulate cortex from September to October 2021. The analyses of Bland-Altman, Pearson correlation and reasonability were performed to assess the degree of agreement, linear correlation and reasonability between the two quantification methods. Results: Fifteen healthy volunteers (12 females and 3 males, age range: 21-35 years, mean age/standard deviation = 27.4/3.9 years) were recruited. The analyses of Bland-Altman, Pearson correlation and reasonability showed high to good consistency and very strong to moderate correlation between the two methods for quantification of total N-acetylaspartate (tNAA), total choline (tCho), and inositol (Ins) (relative half interval of limits of agreement = 3.04%, 9.3%, and 18.5%, respectively; Pearson correlation coefficient r = 0.775, 0.927, and 0.469, respectively). In addition, quantification results of QNet are more likely to be closer to the previous reported average values than those of LCModel. Conclusion: There were high or good degrees of consistency between the quantification results of QNet and LCModel for tNAA, tCho, and Ins, and QNet generally has more reasonable quantification than LCModel.
Distance and Hop-wise Structures Encoding Enhanced Graph Attention Networks
Dec 06, 2021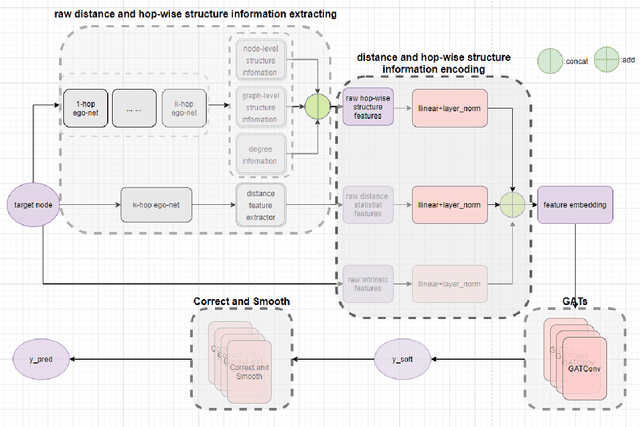
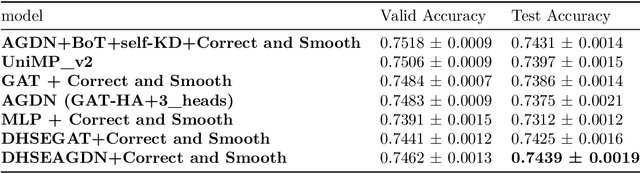
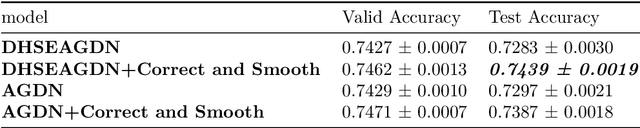
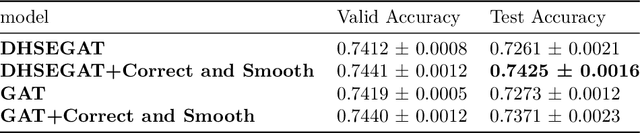
Abstract:Numerous works have proven that existing neighbor-averaging Graph Neural Networks cannot efficiently catch structure features, and many works show that injecting structure, distance, position or spatial features can significantly improve performance of GNNs, however, injecting overall structure and distance into GNNs is an intuitive but remaining untouched idea. In this work, we shed light on the direction. We first extracting hop-wise structure information and compute distance distributional information, gathering with node's intrinsic features, embedding them into same vector space and then adding them up. The derived embedding vectors are then fed into GATs(like GAT, AGDN) and then Correct and Smooth, experiments show that the DHSEGATs achieve competitive result. The code is available at https://github.com/hzg0601/DHSEGATs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge