Yu Mao
Do Models Hear Like Us? Probing the Representational Alignment of Audio LLMs and Naturalistic EEG
Jan 23, 2026Abstract:Audio Large Language Models (Audio LLMs) have demonstrated strong capabilities in integrating speech perception with language understanding. However, whether their internal representations align with human neural dynamics during naturalistic listening remains largely unexplored. In this work, we systematically examine layer-wise representational alignment between 12 open-source Audio LLMs and Electroencephalogram (EEG) signals across 2 datasets. Specifically, we employ 8 similarity metrics, such as Spearman-based Representational Similarity Analysis (RSA), to characterize within-sentence representational geometry. Our analysis reveals 3 key findings: (1) we observe a rank-dependence split, in which model rankings vary substantially across different similarity metrics; (2) we identify spatio-temporal alignment patterns characterized by depth-dependent alignment peaks and a pronounced increase in RSA within the 250-500 ms time window, consistent with N400-related neural dynamics; (3) we find an affective dissociation whereby negative prosody, identified using a proposed Tri-modal Neighborhood Consistency (TNC) criterion, reduces geometric similarity while enhancing covariance-based dependence. These findings provide new neurobiological insights into the representational mechanisms of Audio LLMs.
Lossless Compression of Large Language Model-Generated Text via Next-Token Prediction
May 07, 2025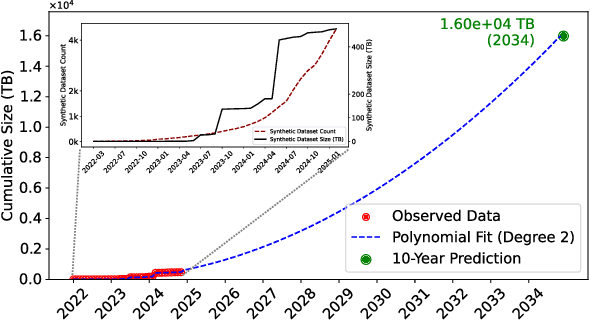
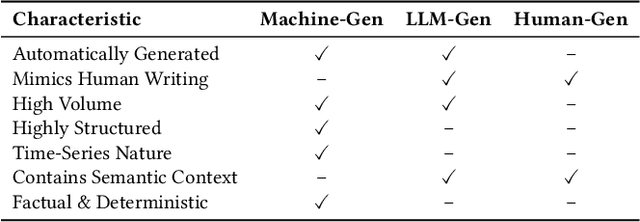
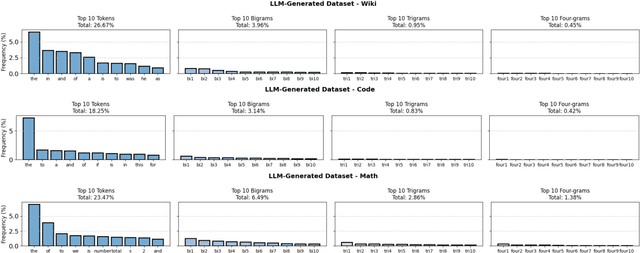
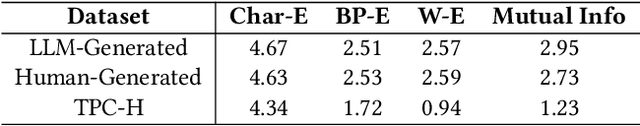
Abstract:As large language models (LLMs) continue to be deployed and utilized across domains, the volume of LLM-generated data is growing rapidly. This trend highlights the increasing importance of effective and lossless compression for such data in modern text management systems. However, compressing LLM-generated data presents unique challenges compared to traditional human- or machine-generated content. Traditional machine-generated data is typically derived from computational processes or device outputs, often highly structured and limited to low-level elements like labels or numerical values. This structure enables conventional lossless compressors to perform efficiently. In contrast, LLM-generated data is more complex and diverse, requiring new approaches for effective compression. In this work, we conduct the first systematic investigation of lossless compression techniques tailored specifically to LLM-generated data. Notably, because LLMs are trained via next-token prediction, we find that LLM-generated data is highly predictable for the models themselves. This predictability enables LLMs to serve as efficient compressors of their own outputs. Through extensive experiments with 14 representative LLMs and 8 LLM-generated datasets from diverse domains, we show that LLM-based prediction methods achieve remarkable compression rates, exceeding 20x, far surpassing the 3x rate achieved by Gzip, a widely used general-purpose compressor. Furthermore, this advantage holds across different LLM sizes and dataset types, demonstrating the robustness and practicality of LLM-based methods in lossless text compression under generative AI workloads.
Easz: An Agile Transformer-based Image Compression Framework for Resource-constrained IoTs
May 03, 2025Abstract:Neural image compression, necessary in various machine-to-machine communication scenarios, suffers from its heavy encode-decode structures and inflexibility in switching between different compression levels. Consequently, it raises significant challenges in applying the neural image compression to edge devices that are developed for powerful servers with high computational and storage capacities. We take a step to solve the challenges by proposing a new transformer-based edge-compute-free image coding framework called Easz. Easz shifts the computational overhead to the server, and hence avoids the heavy encoding and model switching overhead on the edge. Easz utilizes a patch-erase algorithm to selectively remove image contents using a conditional uniform-based sampler. The erased pixels are reconstructed on the receiver side through a transformer-based framework. To further reduce the computational overhead on the receiver, we then introduce a lightweight transformer-based reconstruction structure to reduce the reconstruction load on the receiver side. Extensive evaluations conducted on a real-world testbed demonstrate multiple advantages of Easz over existing compression approaches, in terms of adaptability to different compression levels, computational efficiency, and image reconstruction quality.
When Compression Meets Model Compression: Memory-Efficient Double Compression for Large Language Models
Feb 21, 2025Abstract:Large language models (LLMs) exhibit excellent performance in various tasks. However, the memory requirements of LLMs present a great challenge when deploying on memory-limited devices, even for quantized LLMs. This paper introduces a framework to compress LLM after quantization further, achieving about 2.2x compression ratio. A compression-aware quantization is first proposed to enhance model weight compressibility by re-scaling the model parameters before quantization, followed by a pruning method to improve further. Upon this, we notice that decompression can be a bottleneck during practical scenarios. We then give a detailed analysis of the trade-off between memory usage and latency brought by the proposed method. A speed-adaptive method is proposed to overcome it. The experimental results show inference with the compressed model can achieve a 40% reduction in memory size with negligible loss in accuracy and inference speed.
Turbulence stabilization
Nov 05, 2024Abstract:We recently developed a new approach to get a stabilized image from a sequence of frames acquired through atmospheric turbulence. The goal of this algorihtm is to remove the geometric distortions due by the atmosphere movements. This method is based on a variational formulation and is efficiently solved by the use of Bregman iterations and the operator splitting method. In this paper we propose to study the influence of the choice of the regularizing term in the model. Then we proposed to experiment some of the most used regularization constraints available in the litterature.
Non rigid geometric distortions correction -- Application to atmospheric turbulence stabilization
Nov 04, 2024Abstract:A novel approach is presented to recover an image degraded by atmospheric turbulence. Given a sequence of frames affected by turbulence, we construct a variational model to characterize the static image. The optimization problem is solved by Bregman Iteration and the operator splitting method. Our algorithm is simple, efficient, and can be easily generalized for different scenarios.
SHAP-CAT: A interpretable multi-modal framework enhancing WSI classification via virtual staining and shapley-value-based multimodal fusion
Oct 02, 2024Abstract:The multimodal model has demonstrated promise in histopathology. However, most multimodal models are based on H\&E and genomics, adopting increasingly complex yet black-box designs. In our paper, we propose a novel interpretable multimodal framework named SHAP-CAT, which uses a Shapley-value-based dimension reduction technique for effective multimodal fusion. Starting with two paired modalities -- H\&E and IHC images, we employ virtual staining techniques to enhance limited input data by generating a new clinical-related modality. Lightweight bag-level representations are extracted from image modalities and a Shapley-value-based mechanism is used for dimension reduction. For each dimension of the bag-level representation, attribution values are calculated to indicate how changes in the specific dimensions of the input affect the model output. In this way, we select a few top important dimensions of bag-level representation for each image modality to late fusion. Our experimental results demonstrate that the proposed SHAP-CAT framework incorporating synthetic modalities significantly enhances model performance, yielding a 5\% increase in accuracy for the BCI, an 8\% increase for IHC4BC-ER, and an 11\% increase for the IHC4BC-PR dataset.
Advances in Multiple Instance Learning for Whole Slide Image Analysis: Techniques, Challenges, and Future Directions
Aug 18, 2024



Abstract:Whole slide images (WSIs) are gigapixel-scale digital images of H\&E-stained tissue samples widely used in pathology. The substantial size and complexity of WSIs pose unique analytical challenges. Multiple Instance Learning (MIL) has emerged as a powerful approach for addressing these challenges, particularly in cancer classification and detection. This survey provides a comprehensive overview of the challenges and methodologies associated with applying MIL to WSI analysis, including attention mechanisms, pseudo-labeling, transformers, pooling functions, and graph neural networks. Additionally, it explores the potential of MIL in discovering cancer cell morphology, constructing interpretable machine learning models, and quantifying cancer grading. By summarizing the current challenges, methodologies, and potential applications of MIL in WSI analysis, this survey aims to inform researchers about the state of the field and inspire future research directions.
IHC Matters: Incorporating IHC analysis to H&E Whole Slide Image Analysis for Improved Cancer Grading via Two-stage Multimodal Bilinear Pooling Fusion
May 13, 2024Abstract:Immunohistochemistry (IHC) plays a crucial role in pathology as it detects the over-expression of protein in tissue samples. However, there are still fewer machine learning model studies on IHC's impact on accurate cancer grading. We discovered that IHC and H\&E possess distinct advantages and disadvantages while possessing certain complementary qualities. Building on this observation, we developed a two-stage multi-modal bilinear model with a feature pooling module. This model aims to maximize the potential of both IHC and HE's feature representation, resulting in improved performance compared to their individual use. Our experiments demonstrate that incorporating IHC data into machine learning models, alongside H\&E stained images, leads to superior predictive results for cancer grading. The proposed framework achieves an impressive ACC higher of 0.953 on the public dataset BCI.
Pre-processing matters: A segment search method for WSI classification
Apr 17, 2024



Abstract:Pre-processing for whole slide images can affect classification performance both in the training and inference stages. Our study analyzes the impact of pre-processing parameters on inference and training across single- and multiple-domain datasets. However, searching for an optimal parameter set is time-consuming. To overcome this, we propose a novel Similarity-based Simulated Annealing approach for fast parameter tuning to enhance inference performance on single-domain data. Our method demonstrates significant performance improvements in accuracy, which raise accuracy from 0.512 to 0.847 in a single domain. We further extend our insight into training performance in multi-domain data by employing a novel Bayesian optimization to search optimal pre-processing parameters, resulting in a high AUC of 0.967. We highlight that better pre-processing for WSI can contribute to further accuracy improvement in the histology area.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge