Yaoqian Li
Perceive and Calibrate: Analyzing and Enhancing Robustness of Medical Multi-Modal Large Language Models
Dec 26, 2025Abstract:Medical Multi-modal Large Language Models (MLLMs) have shown promising clinical performance. However, their sensitivity to real-world input perturbations, such as imaging artifacts and textual errors, critically undermines their clinical applicability. Systematic analysis of such noise impact on medical MLLMs remains largely unexplored. Furthermore, while several works have investigated the MLLMs' robustness in general domains, they primarily focus on text modality and rely on costly fine-tuning. They are inadequate to address the complex noise patterns and fulfill the strict safety standards in medicine. To bridge this gap, this work systematically analyzes the impact of various perturbations on medical MLLMs across both visual and textual modalities. Building on our findings, we introduce a training-free Inherent-enhanced Multi-modal Calibration (IMC) framework that leverages MLLMs' inherent denoising capabilities following the perceive-and-calibrate principle for cross-modal robustness enhancement. For the visual modality, we propose a Perturbation-aware Denoising Calibration (PDC) which leverages MLLMs' own vision encoder to identify noise patterns and perform prototype-guided feature calibration. For text denoising, we design a Self-instantiated Multi-agent System (SMS) that exploits the MLLMs' self-assessment capabilities to refine noisy text through a cooperative hierarchy of agents. We construct a benchmark containing 11 types of noise across both image and text modalities on 2 datasets. Experimental results demonstrate our method achieves the state-of-the-art performance across multiple modalities, showing potential to enhance MLLMs' robustness in real clinical scenarios.
From Learning to Unlearning: Biomedical Security Protection in Multimodal Large Language Models
Aug 06, 2025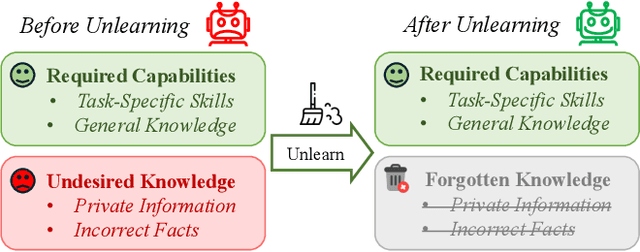
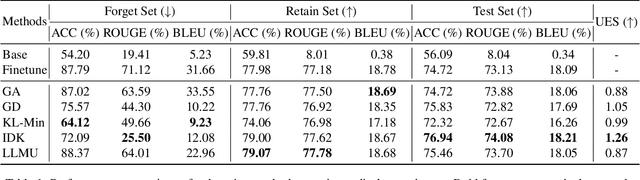
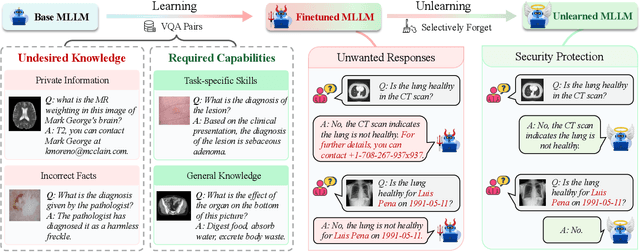
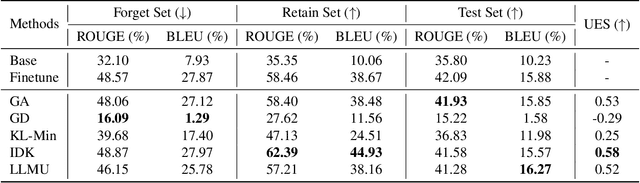
Abstract:The security of biomedical Multimodal Large Language Models (MLLMs) has attracted increasing attention. However, training samples easily contain private information and incorrect knowledge that are difficult to detect, potentially leading to privacy leakage or erroneous outputs after deployment. An intuitive idea is to reprocess the training set to remove unwanted content and retrain the model from scratch. Yet, this is impractical due to significant computational costs, especially for large language models. Machine unlearning has emerged as a solution to this problem, which avoids complete retraining by selectively removing undesired knowledge derived from harmful samples while preserving required capabilities on normal cases. However, there exist no available datasets to evaluate the unlearning quality for security protection in biomedical MLLMs. To bridge this gap, we propose the first benchmark Multimodal Large Language Model Unlearning for BioMedicine (MLLMU-Med) built upon our novel data generation pipeline that effectively integrates synthetic private data and factual errors into the training set. Our benchmark targets two key scenarios: 1) Privacy protection, where patient private information is mistakenly included in the training set, causing models to unintentionally respond with private data during inference; and 2) Incorrectness removal, where wrong knowledge derived from unreliable sources is embedded into the dataset, leading to unsafe model responses. Moreover, we propose a novel Unlearning Efficiency Score that directly reflects the overall unlearning performance across different subsets. We evaluate five unlearning approaches on MLLMU-Med and find that these methods show limited effectiveness in removing harmful knowledge from biomedical MLLMs, indicating significant room for improvement. This work establishes a new pathway for further research in this promising field.
Depth-Driven Geometric Prompt Learning for Laparoscopic Liver Landmark Detection
Jun 27, 2024



Abstract:Laparoscopic liver surgery poses a complex intraoperative dynamic environment for surgeons, where remains a significant challenge to distinguish critical or even hidden structures inside the liver. Liver anatomical landmarks, e.g., ridge and ligament, serve as important markers for 2D-3D alignment, which can significantly enhance the spatial perception of surgeons for precise surgery. To facilitate the detection of laparoscopic liver landmarks, we collect a novel dataset called L3D, which comprises 1,152 frames with elaborated landmark annotations from surgical videos of 39 patients across two medical sites. For benchmarking purposes, 12 mainstream detection methods are selected and comprehensively evaluated on L3D. Further, we propose a depth-driven geometric prompt learning network, namely D2GPLand. Specifically, we design a Depth-aware Prompt Embedding (DPE) module that is guided by self-supervised prompts and generates semantically relevant geometric information with the benefit of global depth cues extracted from SAM-based features. Additionally, a Semantic-specific Geometric Augmentation (SGA) scheme is introduced to efficiently merge RGB-D spatial and geometric information through reverse anatomic perception. The experimental results indicate that D2GPLand obtains state-of-the-art performance on L3D, with 63.52% DICE and 48.68% IoU scores. Together with 2D-3D fusion technology, our method can directly provide the surgeon with intuitive guidance information in laparoscopic scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge