Dunyuan Xu
From Learning to Unlearning: Biomedical Security Protection in Multimodal Large Language Models
Aug 06, 2025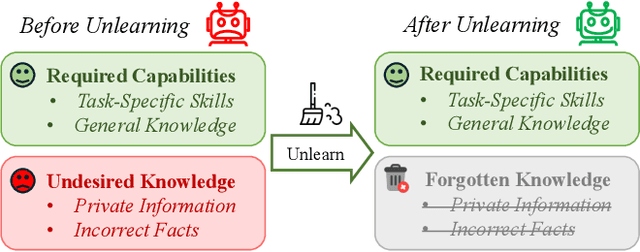
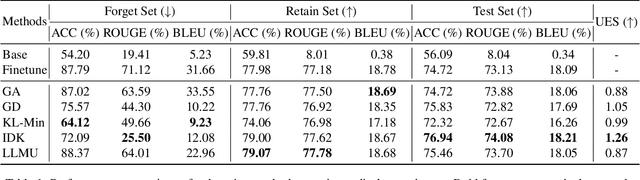
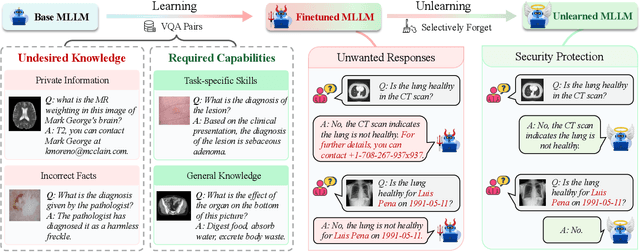
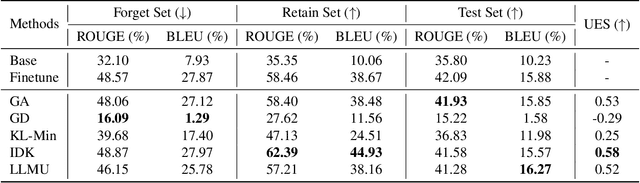
Abstract:The security of biomedical Multimodal Large Language Models (MLLMs) has attracted increasing attention. However, training samples easily contain private information and incorrect knowledge that are difficult to detect, potentially leading to privacy leakage or erroneous outputs after deployment. An intuitive idea is to reprocess the training set to remove unwanted content and retrain the model from scratch. Yet, this is impractical due to significant computational costs, especially for large language models. Machine unlearning has emerged as a solution to this problem, which avoids complete retraining by selectively removing undesired knowledge derived from harmful samples while preserving required capabilities on normal cases. However, there exist no available datasets to evaluate the unlearning quality for security protection in biomedical MLLMs. To bridge this gap, we propose the first benchmark Multimodal Large Language Model Unlearning for BioMedicine (MLLMU-Med) built upon our novel data generation pipeline that effectively integrates synthetic private data and factual errors into the training set. Our benchmark targets two key scenarios: 1) Privacy protection, where patient private information is mistakenly included in the training set, causing models to unintentionally respond with private data during inference; and 2) Incorrectness removal, where wrong knowledge derived from unreliable sources is embedded into the dataset, leading to unsafe model responses. Moreover, we propose a novel Unlearning Efficiency Score that directly reflects the overall unlearning performance across different subsets. We evaluate five unlearning approaches on MLLMU-Med and find that these methods show limited effectiveness in removing harmful knowledge from biomedical MLLMs, indicating significant room for improvement. This work establishes a new pathway for further research in this promising field.
Towards Synchronous Memorizability and Generalizability with Site-Modulated Diffusion Replay for Cross-Site Continual Segmentation
Jun 26, 2024Abstract:The ability to learn sequentially from different data sites is crucial for a deep network in solving practical medical image diagnosis problems due to privacy restrictions and storage limitations. However, adapting on incoming site leads to catastrophic forgetting on past sites and decreases generalizablity on unseen sites. Existing Continual Learning (CL) and Domain Generalization (DG) methods have been proposed to solve these two challenges respectively, but none of them can address both simultaneously. Recognizing this limitation, this paper proposes a novel training paradigm, learning towards Synchronous Memorizability and Generalizability (SMG-Learning). To achieve this, we create the orientational gradient alignment to ensure memorizability on previous sites, and arbitrary gradient alignment to enhance generalizability on unseen sites. This approach is named as Parallel Gradient Alignment (PGA). Furthermore, we approximate the PGA as dual meta-objectives using the first-order Taylor expansion to reduce computational cost of aligning gradients. Considering that performing gradient alignments, especially for previous sites, is not feasible due to the privacy constraints, we design a Site-Modulated Diffusion (SMD) model to generate images with site-specific learnable prompts, replaying images have similar data distributions as previous sites. We evaluate our method on two medical image segmentation tasks, where data from different sites arrive sequentially. Experimental results show that our method efficiently enhances both memorizability and generalizablity better than other state-of-the-art methods, delivering satisfactory performance across all sites. Our code will be available at: https://github.com/dyxu-cuhkcse/SMG-Learning.
Cross-modality Guidance-aided Multi-modal Learning with Dual Attention for MRI Brain Tumor Grading
Jan 17, 2024Abstract:Brain tumor represents one of the most fatal cancers around the world, and is very common in children and the elderly. Accurate identification of the type and grade of tumor in the early stages plays an important role in choosing a precise treatment plan. The Magnetic Resonance Imaging (MRI) protocols of different sequences provide clinicians with important contradictory information to identify tumor regions. However, manual assessment is time-consuming and error-prone due to big amount of data and the diversity of brain tumor types. Hence, there is an unmet need for MRI automated brain tumor diagnosis. We observe that the predictive capability of uni-modality models is limited and their performance varies widely across modalities, and the commonly used modality fusion methods would introduce potential noise, which results in significant performance degradation. To overcome these challenges, we propose a novel cross-modality guidance-aided multi-modal learning with dual attention for addressing the task of MRI brain tumor grading. To balance the tradeoff between model efficiency and efficacy, we employ ResNet Mix Convolution as the backbone network for feature extraction. Besides, dual attention is applied to capture the semantic interdependencies in spatial and slice dimensions respectively. To facilitate information interaction among modalities, we design a cross-modality guidance-aided module where the primary modality guides the other secondary modalities during the process of training, which can effectively leverage the complementary information of different MRI modalities and meanwhile alleviate the impact of the possible noise.
Pseudo Bias-Balanced Learning for Debiased Chest X-ray Classification
Mar 18, 2022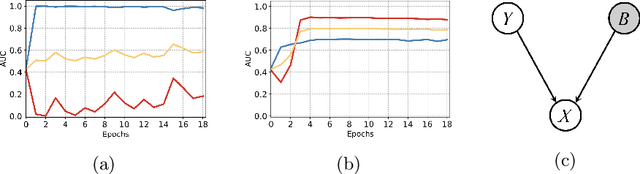
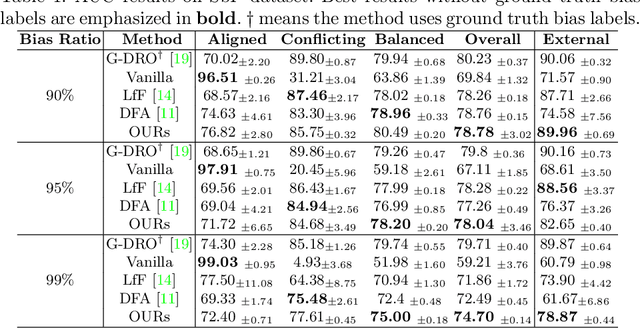
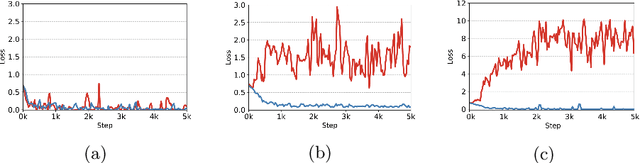
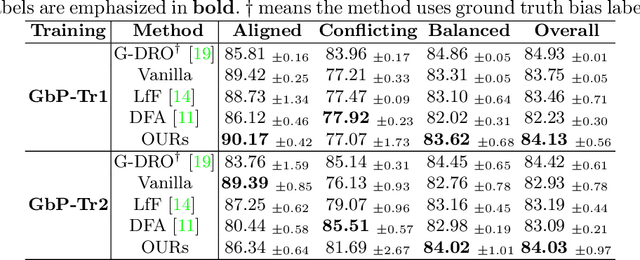
Abstract:Deep learning models were frequently reported to learn from shortcuts like dataset biases. As deep learning is playing an increasingly important role in the modern healthcare system, it is of great need to combat shortcut learning in medical data as well as develop unbiased and trustworthy models. In this paper, we study the problem of developing debiased chest X-ray diagnosis models from the biased training data without knowing exactly the bias labels. We start with the observations that the imbalance of bias distribution is one of the key reasons causing shortcut learning, and the dataset biases are preferred by the model if they were easier to be learned than the intended features. Based on these observations, we propose a novel algorithm, pseudo bias-balanced learning, which first captures and predicts per-sample bias labels via generalized cross entropy loss and then trains a debiased model using pseudo bias labels and bias-balanced softmax function. To our best knowledge, we are pioneered in tackling dataset biases in medical images without explicit labeling on the bias attributes. We constructed several chest X-ray datasets with various dataset bias situations and demonstrated with extensive experiments that our proposed method achieved consistent improvements over other state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge