Xuan Xu
Fanny
HySim-LLM: Embedding-Weighted Fine-Tuning Bounds and Manifold Denoising for Domain-Adapted LLMs
Oct 09, 2025Abstract:The extraction and standardization of pharmacokinetic (PK) information from scientific literature remain significant challenges in computational pharmacology, which limits the reliability of data-driven models in drug development. Large language models (LLMs) have achieved remarkable progress in text understanding and reasoning, yet their adaptation to structured biomedical data, such as PK tables, remains constrained by heterogeneity, noise, and domain shift. To address these limitations, we propose HySim-LLM, a unified mathematical and computational framework that integrates embedding-weighted fine-tuning and manifold-aware denoising to enhance the robustness and interpretability of LLMs. We establish two theoretical results: (1) a similarity-weighted generalization bound that quantifies adaptation performance under embedding divergence, and (2) a manifold-based denoising guarantee that bounds loss contributions from noisy or off-manifold samples. These theorems provide a principled foundation for fine-tuning LLMs in structured biomedical settings. The framework offers a mathematically grounded pathway toward reliable and interpretable LLM adaptation for biomedical and data-intensive scientific domains.
Predictive Modeling and Explainable AI for Veterinary Safety Profiles, Residue Assessment, and Health Outcomes Using Real-World Data and Physicochemical Properties
Oct 01, 2025Abstract:The safe use of pharmaceuticals in food-producing animals is vital to protect animal welfare and human food safety. Adverse events (AEs) may signal unexpected pharmacokinetic or toxicokinetic effects, increasing the risk of violative residues in the food chain. This study introduces a predictive framework for classifying outcomes (Death vs. Recovery) using ~1.28 million reports (1987-2025 Q1) from the U.S. FDA's OpenFDA Center for Veterinary Medicine. A preprocessing pipeline merged relational tables and standardized AEs through VeDDRA ontologies. Data were normalized, missing values imputed, and high-cardinality features reduced; physicochemical drug properties were integrated to capture chemical-residue links. We evaluated supervised models, including Random Forest, CatBoost, XGBoost, ExcelFormer, and large language models (Gemma 3-27B, Phi 3-12B). Class imbalance was addressed, such as undersampling and oversampling, with a focus on prioritizing recall for fatal outcomes. Ensemble methods(Voting, Stacking) and CatBoost performed best, achieving precision, recall, and F1-scores of 0.95. Incorporating Average Uncertainty Margin (AUM)-based pseudo-labeling of uncertain cases improved minority-class detection, particularly in ExcelFormer and XGBoost. Interpretability via SHAP identified biologically plausible predictors, including lung, heart, and bronchial disorders, animal demographics, and drug physicochemical properties. These features were strongly linked to fatal outcomes. Overall, the framework shows that combining rigorous data engineering, advanced machine learning, and explainable AI enables accurate, interpretable predictions of veterinary safety outcomes. The approach supports FARAD's mission by enabling early detection of high-risk drug-event profiles, strengthening residue risk assessment, and informing regulatory and clinical decision-making.
Reduced Spatial Dependency for More General Video-level Deepfake Detection
Mar 05, 2025



Abstract:As one of the prominent AI-generated content, Deepfake has raised significant safety concerns. Although it has been demonstrated that temporal consistency cues offer better generalization capability, existing methods based on CNNs inevitably introduce spatial bias, which hinders the extraction of intrinsic temporal features. To address this issue, we propose a novel method called Spatial Dependency Reduction (SDR), which integrates common temporal consistency features from multiple spatially-perturbed clusters, to reduce the dependency of the model on spatial information. Specifically, we design multiple Spatial Perturbation Branch (SPB) to construct spatially-perturbed feature clusters. Subsequently, we utilize the theory of mutual information and propose a Task-Relevant Feature Integration (TRFI) module to capture temporal features residing in similar latent space from these clusters. Finally, the integrated feature is fed into a temporal transformer to capture long-range dependencies. Extensive benchmarks and ablation studies demonstrate the effectiveness and rationale of our approach.
FIRE: Robust Detection of Diffusion-Generated Images via Frequency-Guided Reconstruction Error
Dec 10, 2024



Abstract:The rapid advancement of diffusion models has significantly improved high-quality image generation, making generated content increasingly challenging to distinguish from real images and raising concerns about potential misuse. In this paper, we observe that diffusion models struggle to accurately reconstruct mid-band frequency information in real images, suggesting the limitation could serve as a cue for detecting diffusion model generated images. Motivated by this observation, we propose a novel method called Frequency-guided Reconstruction Error (FIRE), which, to the best of our knowledge, is the first to investigate the influence of frequency decomposition on reconstruction error. FIRE assesses the variation in reconstruction error before and after the frequency decomposition, offering a robust method for identifying diffusion model generated images. Extensive experiments show that FIRE generalizes effectively to unseen diffusion models and maintains robustness against diverse perturbations.
Histo-Diffusion: A Diffusion Super-Resolution Method for Digital Pathology with Comprehensive Quality Assessment
Aug 27, 2024



Abstract:Digital pathology has advanced significantly over the last decade, with Whole Slide Images (WSIs) encompassing vast amounts of data essential for accurate disease diagnosis. High-resolution WSIs are essential for precise diagnosis but technical limitations in scanning equipment and variablity in slide preparation can hinder obtaining these images. Super-resolution techniques can enhance low-resolution images; while Generative Adversarial Networks (GANs) have been effective in natural image super-resolution tasks, they often struggle with histopathology due to overfitting and mode collapse. Traditional evaluation metrics fall short in assessing the complex characteristics of histopathology images, necessitating robust histology-specific evaluation methods. We introduce Histo-Diffusion, a novel diffusion-based method specially designed for generating and evaluating super-resolution images in digital pathology. It includes a restoration module for histopathology prior and a controllable diffusion module for generating high-quality images. We have curated two histopathology datasets and proposed a comprehensive evaluation strategy which incorporates both full-reference and no-reference metrics to thoroughly assess the quality of digital pathology images. Comparative analyses on multiple datasets with state-of-the-art methods reveal that Histo-Diffusion outperforms GANs. Our method offers a versatile solution for histopathology image super-resolution, capable of handling multi-resolution generation from varied input sizes, providing valuable support in diagnostic processes.
Unearthing Common Inconsistency for Generalisable Deepfake Detection
Nov 20, 2023



Abstract:Deepfake has emerged for several years, yet efficient detection techniques could generalize over different manipulation methods require further research. While current image-level detection method fails to generalize to unseen domains, owing to the domain-shift phenomenon brought by CNN's strong inductive bias towards Deepfake texture, video-level one shows its potential to have both generalization across multiple domains and robustness to compression. We argue that although distinct face manipulation tools have different inherent bias, they all disrupt the consistency between frames, which is a natural characteristic shared by authentic videos. Inspired by this, we proposed a detection approach by capturing frame inconsistency that broadly exists in different forgery techniques, termed unearthing-common-inconsistency (UCI). Concretely, the UCI network based on self-supervised contrastive learning can better distinguish temporal consistency between real and fake videos from multiple domains. We introduced a temporally-preserved module method to introduce spatial noise perturbations, directing the model's attention towards temporal information. Subsequently, leveraging a multi-view cross-correlation learning module, we extensively learn the disparities in temporal representations between genuine and fake samples. Extensive experiments demonstrate the generalization ability of our method on unseen Deepfake domains.
ViT-DAE: Transformer-driven Diffusion Autoencoder for Histopathology Image Analysis
Apr 03, 2023Abstract:Generative AI has received substantial attention in recent years due to its ability to synthesize data that closely resembles the original data source. While Generative Adversarial Networks (GANs) have provided innovative approaches for histopathological image analysis, they suffer from limitations such as mode collapse and overfitting in discriminator. Recently, Denoising Diffusion models have demonstrated promising results in computer vision. These models exhibit superior stability during training, better distribution coverage, and produce high-quality diverse images. Additionally, they display a high degree of resilience to noise and perturbations, making them well-suited for use in digital pathology, where images commonly contain artifacts and exhibit significant variations in staining. In this paper, we present a novel approach, namely ViT-DAE, which integrates vision transformers (ViT) and diffusion autoencoders for high-quality histopathology image synthesis. This marks the first time that ViT has been introduced to diffusion autoencoders in computational pathology, allowing the model to better capture the complex and intricate details of histopathology images. We demonstrate the effectiveness of ViT-DAE on three publicly available datasets. Our approach outperforms recent GAN-based and vanilla DAE methods in generating realistic images.
Enhancing Modality-Agnostic Representations via Meta-Learning for Brain Tumor Segmentation
Feb 08, 2023



Abstract:In the medical vision domain, different imaging modalities provide complementary information. However, in practice, not all modalities may be available during inference. Previous approaches, e.g., knowledge distillation or image synthesis, often assume the availability of full modalities for all patients during training; this is unrealistic and impractical owing to the variability in data collection across sites. We propose a novel approach to learn enhanced modality-agnostic representations by employing a novel meta-learning strategy in training, even when only a fraction of full modality patients are available. Meta-learning enhances partial modality representations to full modality representations by meta-training on partial modality data and meta-testing on limited full modality samples. Additionally, we co-supervise this feature enrichment by introducing an auxiliary adversarial learning branch. More specifically, a missing modality detector is used as a discriminator to mimic the full modality setting. Our segmentation framework significantly outperforms state-of-the-art brain tumor segmentation techniques in missing modality scenarios, as demonstrated on two brain tumor MRI datasets.
Temporal Context Matters: Enhancing Single Image Prediction with Disease Progression Representations
Mar 31, 2022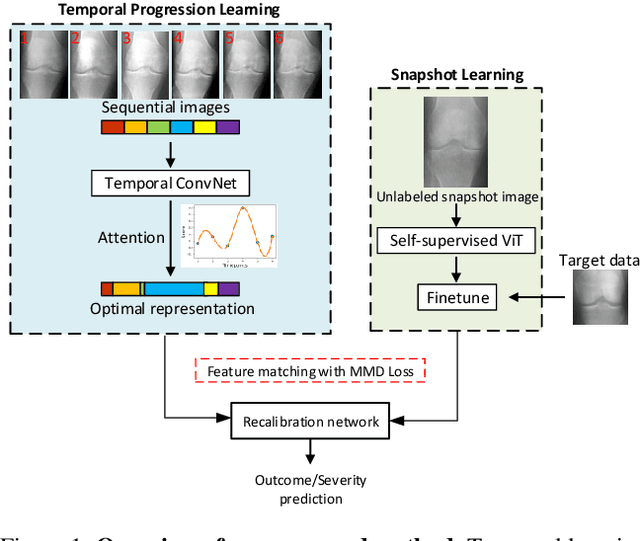
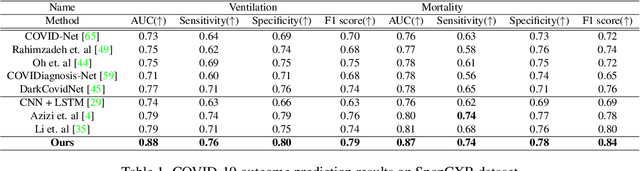
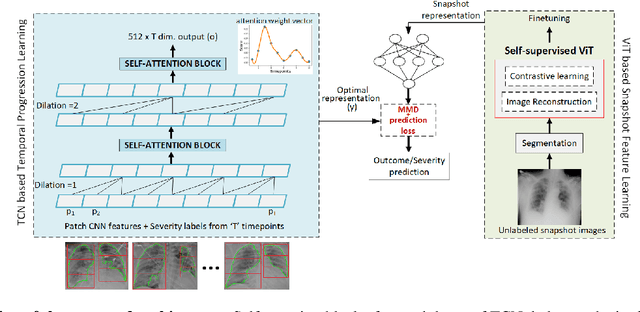
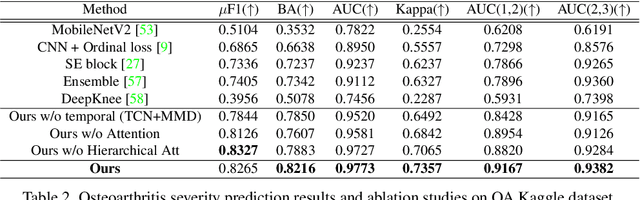
Abstract:Clinical outcome or severity prediction from medical images has largely focused on learning representations from single-timepoint or snapshot scans. It has been shown that disease progression can be better characterized by temporal imaging. We therefore hypothesized that outcome predictions can be improved by utilizing the disease progression information from sequential images. We present a deep learning approach that leverages temporal progression information to improve clinical outcome predictions from single-timepoint images. In our method, a self-attention based Temporal Convolutional Network (TCN) is used to learn a representation that is most reflective of the disease trajectory. Meanwhile, a Vision Transformer is pretrained in a self-supervised fashion to extract features from single-timepoint images. The key contribution is to design a recalibration module that employs maximum mean discrepancy loss (MMD) to align distributions of the above two contextual representations. We train our system to predict clinical outcomes and severity grades from single-timepoint images. Experiments on chest and osteoarthritis radiography datasets demonstrate that our approach outperforms other state-of-the-art techniques.
Brain Cancer Survival Prediction on Treatment-na ive MRI using Deep Anchor Attention Learning with Vision Transformer
Feb 03, 2022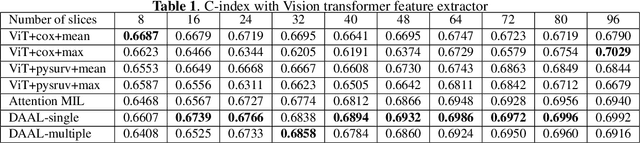
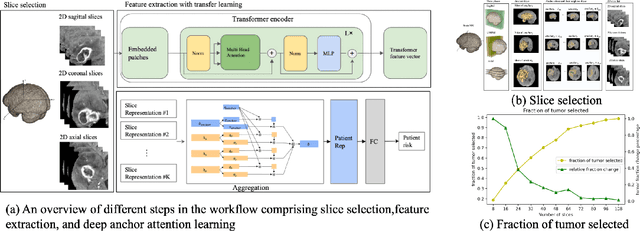
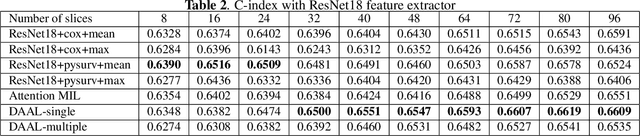
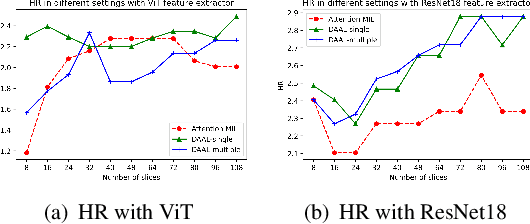
Abstract:Image-based brain cancer prediction models, based on radiomics, quantify the radiologic phenotype from magnetic resonance imaging (MRI). However, these features are difficult to reproduce because of variability in acquisition and preprocessing pipelines. Despite evidence of intra-tumor phenotypic heterogeneity, the spatial diversity between different slices within an MRI scan has been relatively unexplored using such methods. In this work, we propose a deep anchor attention aggregation strategy with a Vision Transformer to predict survival risk for brain cancer patients. A Deep Anchor Attention Learning (DAAL) algorithm is proposed to assign different weights to slice-level representations with trainable distance measurements. We evaluated our method on N = 326 MRIs. Our results outperformed attention multiple instance learning-based techniques. DAAL highlights the importance of critical slices and corroborates the clinical intuition that inter-slice spatial diversity can reflect disease severity and is implicated in outcome.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge