Majid Jaberi-Douraki
HySim-LLM: Embedding-Weighted Fine-Tuning Bounds and Manifold Denoising for Domain-Adapted LLMs
Oct 09, 2025Abstract:The extraction and standardization of pharmacokinetic (PK) information from scientific literature remain significant challenges in computational pharmacology, which limits the reliability of data-driven models in drug development. Large language models (LLMs) have achieved remarkable progress in text understanding and reasoning, yet their adaptation to structured biomedical data, such as PK tables, remains constrained by heterogeneity, noise, and domain shift. To address these limitations, we propose HySim-LLM, a unified mathematical and computational framework that integrates embedding-weighted fine-tuning and manifold-aware denoising to enhance the robustness and interpretability of LLMs. We establish two theoretical results: (1) a similarity-weighted generalization bound that quantifies adaptation performance under embedding divergence, and (2) a manifold-based denoising guarantee that bounds loss contributions from noisy or off-manifold samples. These theorems provide a principled foundation for fine-tuning LLMs in structured biomedical settings. The framework offers a mathematically grounded pathway toward reliable and interpretable LLM adaptation for biomedical and data-intensive scientific domains.
Predictive Modeling and Explainable AI for Veterinary Safety Profiles, Residue Assessment, and Health Outcomes Using Real-World Data and Physicochemical Properties
Oct 01, 2025Abstract:The safe use of pharmaceuticals in food-producing animals is vital to protect animal welfare and human food safety. Adverse events (AEs) may signal unexpected pharmacokinetic or toxicokinetic effects, increasing the risk of violative residues in the food chain. This study introduces a predictive framework for classifying outcomes (Death vs. Recovery) using ~1.28 million reports (1987-2025 Q1) from the U.S. FDA's OpenFDA Center for Veterinary Medicine. A preprocessing pipeline merged relational tables and standardized AEs through VeDDRA ontologies. Data were normalized, missing values imputed, and high-cardinality features reduced; physicochemical drug properties were integrated to capture chemical-residue links. We evaluated supervised models, including Random Forest, CatBoost, XGBoost, ExcelFormer, and large language models (Gemma 3-27B, Phi 3-12B). Class imbalance was addressed, such as undersampling and oversampling, with a focus on prioritizing recall for fatal outcomes. Ensemble methods(Voting, Stacking) and CatBoost performed best, achieving precision, recall, and F1-scores of 0.95. Incorporating Average Uncertainty Margin (AUM)-based pseudo-labeling of uncertain cases improved minority-class detection, particularly in ExcelFormer and XGBoost. Interpretability via SHAP identified biologically plausible predictors, including lung, heart, and bronchial disorders, animal demographics, and drug physicochemical properties. These features were strongly linked to fatal outcomes. Overall, the framework shows that combining rigorous data engineering, advanced machine learning, and explainable AI enables accurate, interpretable predictions of veterinary safety outcomes. The approach supports FARAD's mission by enabling early detection of high-risk drug-event profiles, strengthening residue risk assessment, and informing regulatory and clinical decision-making.
Knowledge in Triples for LLMs: Enhancing Table QA Accuracy with Semantic Extraction
Sep 21, 2024Abstract:Integrating structured knowledge from tabular formats poses significant challenges within natural language processing (NLP), mainly when dealing with complex, semi-structured tables like those found in the FeTaQA dataset. These tables require advanced methods to interpret and generate meaningful responses accurately. Traditional approaches, such as SQL and SPARQL, often fail to fully capture the semantics of such data, especially in the presence of irregular table structures like web tables. This paper addresses these challenges by proposing a novel approach that extracts triples straightforward from tabular data and integrates it with a retrieval-augmented generation (RAG) model to enhance the accuracy, coherence, and contextual richness of responses generated by a fine-tuned GPT-3.5-turbo-0125 model. Our approach significantly outperforms existing baselines on the FeTaQA dataset, particularly excelling in Sacre-BLEU and ROUGE metrics. It effectively generates contextually accurate and detailed long-form answers from tables, showcasing its strength in complex data interpretation.
Large-Scale Data Mining of Rapid Residue Detection Assay Data From HTML and Documents: Improving Data Access and Visualization for Veterinarians
Dec 02, 2021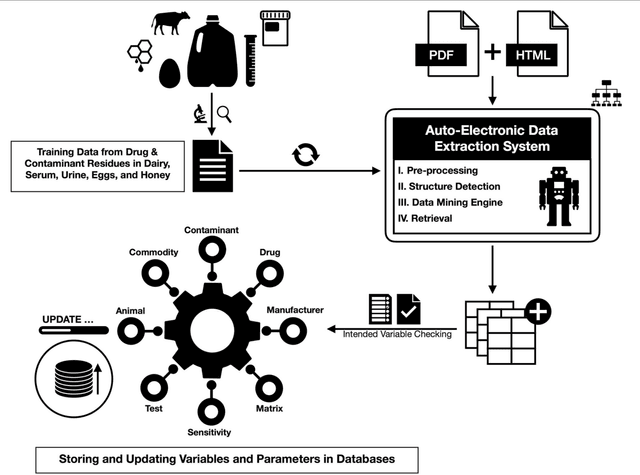

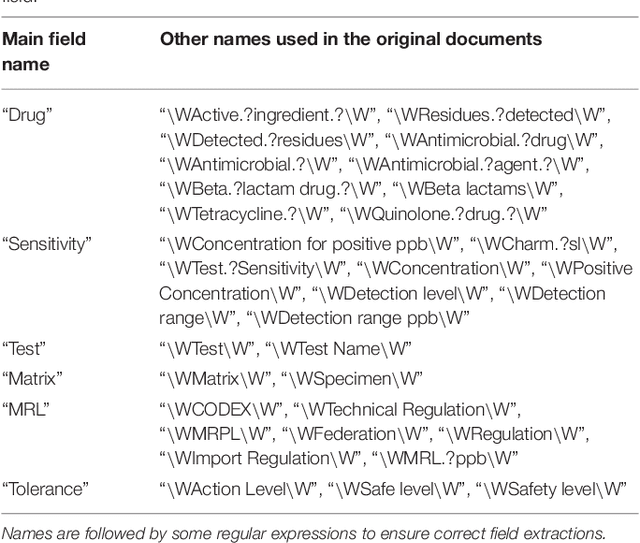
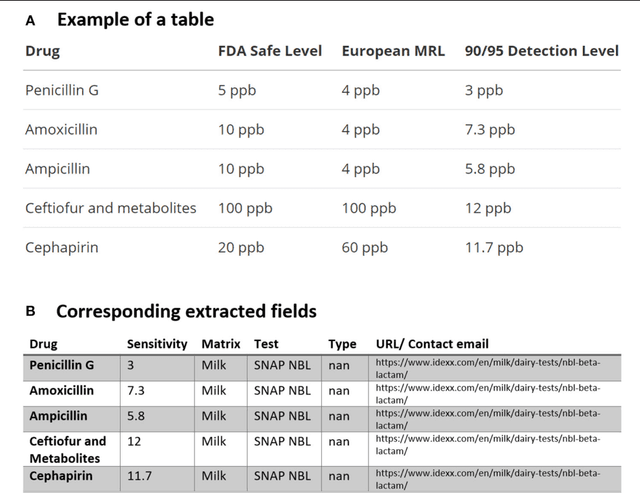
Abstract:Extra-label drug use in food animal medicine is authorized by the US Animal Medicinal Drug Use Clarification Act (AMDUCA), and estimated withdrawal intervals are based on published scientific pharmacokinetic data. Occasionally there is a paucity of scientific data on which to base a withdrawal interval or a large number of animals being treated, driving the need to test for drug residues. Rapid assay commercial farm-side tests are essential for monitoring drug residues in animal products to protect human health. Active ingredients, sensitivity, matrices, and species that have been evaluated for commercial rapid assay tests are typically reported on manufacturers' websites or in PDF documents that are available to consumers but may require a special access request. Additionally, this information is not always correlated with FDA-approved tolerances. Furthermore, parameter changes for these tests can be very challenging to regularly identify, especially those listed on websites or in documents that are not publicly available. Therefore, artificial intelligence plays a critical role in efficiently extracting the data and ensure current information. Extracting tables from PDF and HTML documents has been investigated both by academia and commercial tool builders. Research in text mining of such documents has become a widespread yet challenging arena in implementing natural language programming. However, techniques of extracting tables are still in their infancy and being investigated and improved by researchers. In this study, we developed and evaluated a data-mining method for automatically extracting rapid assay data from electronic documents. Our automatic electronic data extraction method includes a software package module, a developed pattern recognition tool, and a data mining engine. Assay details were provided by several commercial entities that produce these rapid drug residue assay
* 13 pages, 7 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge