Xiangrong Liu
DeepDR: an integrated deep-learning model web server for drug repositioning
Nov 12, 2025Abstract:Background: Identifying new indications for approved drugs is a complex and time-consuming process that requires extensive knowledge of pharmacology, clinical data, and advanced computational methods. Recently, deep learning (DL) methods have shown their capability for the accurate prediction of drug repositioning. However, implementing DL-based modeling requires in-depth domain knowledge and proficient programming skills. Results: In this application, we introduce DeepDR, the first integrated platform that combines a variety of established DL-based models for disease- and target-specific drug repositioning tasks. DeepDR leverages invaluable experience to recommend candidate drugs, which covers more than 15 networks and a comprehensive knowledge graph that includes 5.9 million edges across 107 types of relationships connecting drugs, diseases, proteins/genes, pathways, and expression from six existing databases and a large scientific corpus of 24 million PubMed publications. Additionally, the recommended results include detailed descriptions of the recommended drugs and visualize key patterns with interpretability through a knowledge graph. Conclusion: DeepDR is free and open to all users without the requirement of registration. We believe it can provide an easy-to-use, systematic, highly accurate, and computationally automated platform for both experimental and computational scientists.
LOHA: Direct Graph Spectral Contrastive Learning Between Low-pass and High-pass Views
Jan 06, 2025



Abstract:Spectral Graph Neural Networks effectively handle graphs with different homophily levels, with low-pass filter mining feature smoothness and high-pass filter capturing differences. When these distinct filters could naturally form two opposite views for self-supervised learning, the commonalities between the counterparts for the same node remain unexplored, leading to suboptimal performance. In this paper, a simple yet effective self-supervised contrastive framework, LOHA, is proposed to address this gap. LOHA optimally leverages low-pass and high-pass views by embracing "harmony in diversity". Rather than solely maximizing the difference between these distinct views, which may lead to feature separation, LOHA harmonizes the diversity by treating the propagation of graph signals from both views as a composite feature. Specifically, a novel high-dimensional feature named spectral signal trend is proposed to serve as the basis for the composite feature, which remains relatively unaffected by changing filters and focuses solely on original feature differences. LOHA achieves an average performance improvement of 2.8% over runner-up models on 9 real-world datasets with varying homophily levels. Notably, LOHA even surpasses fully-supervised models on several datasets, which underscores the potential of LOHA in advancing the efficacy of spectral GNNs for diverse graph structures.
Heterogeneous network-based drug repurposing for COVID-19
Jul 20, 2021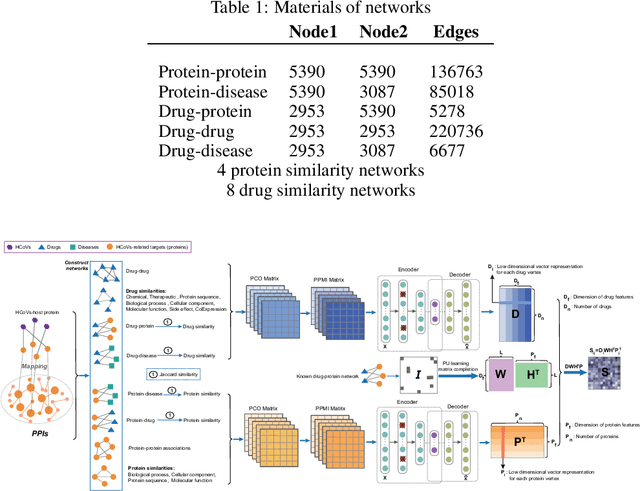
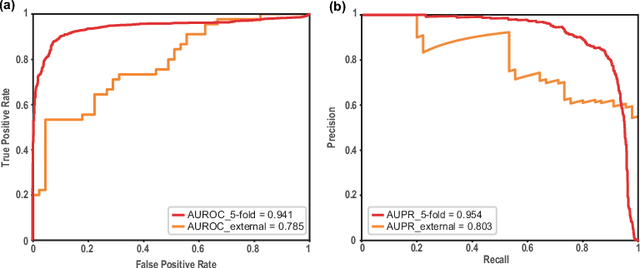
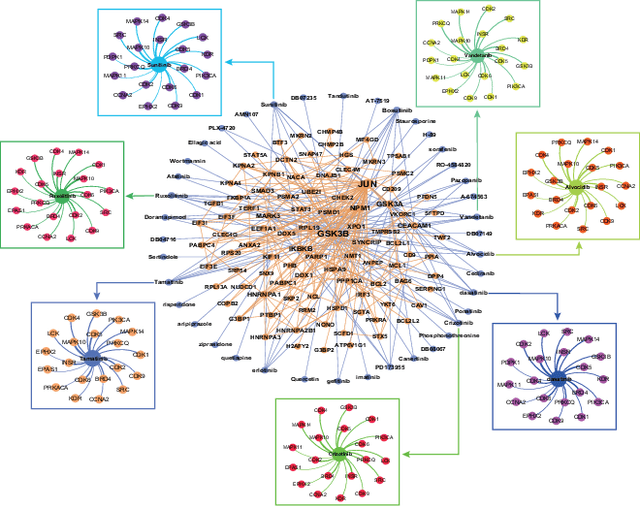
Abstract:The Corona Virus Disease 2019 (COVID-19) belongs to human coronaviruses (HCoVs), which spreads rapidly around the world. Compared with new drug development, drug repurposing may be the best shortcut for treating COVID-19. Therefore, we constructed a comprehensive heterogeneous network based on the HCoVs-related target proteins and use the previously proposed deepDTnet, to discover potential drug candidates for COVID-19. We obtain high performance in predicting the possible drugs effective for COVID-19 related proteins. In summary, this work utilizes a powerful heterogeneous network-based deep learning method, which may be beneficial to quickly identify candidate repurposable drugs toward future clinical trials for COVID-19. The code and data are available at https://github.com/stjin-XMU/HnDR-COVID.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge