Thomas J. Fuchs
JPL
Deep Interactive Learning: An Efficient Labeling Approach for Deep Learning-Based Osteosarcoma Treatment Response Assessment
Jul 02, 2020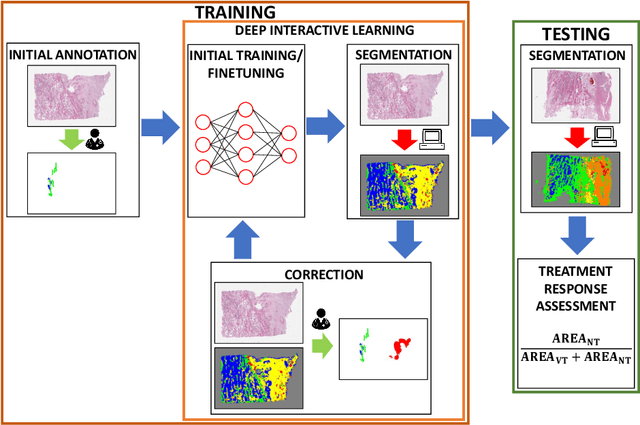
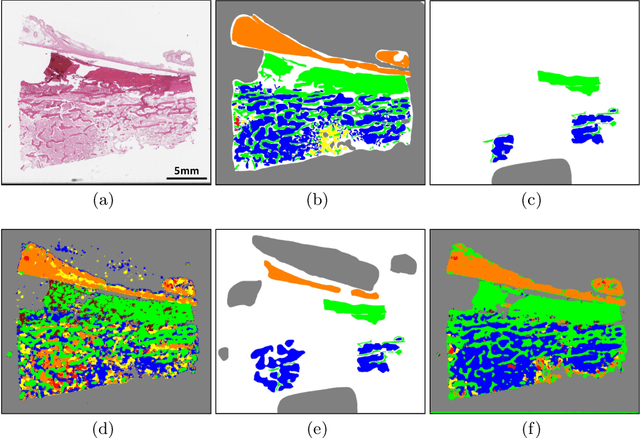
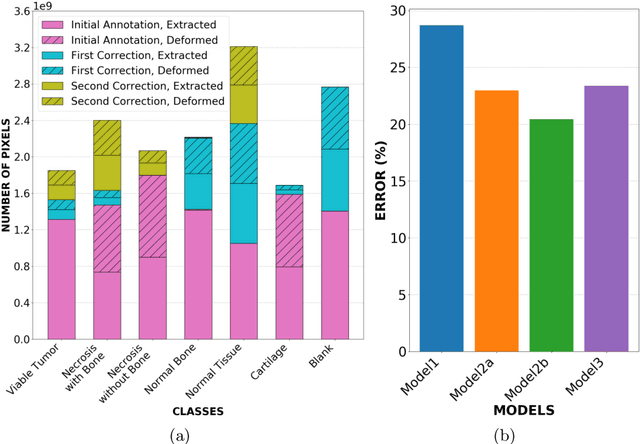
Abstract:Osteosarcoma is the most common malignant primary bone tumor. Standard treatment includes pre-operative chemotherapy followed by surgical resection. The response to treatment as measured by ratio of necrotic tumor area to overall tumor area is a known prognostic factor for overall survival. This assessment is currently done manually by pathologists by looking at glass slides under the microscope which may not be reproducible due to its subjective nature. Convolutional neural networks (CNNs) can be used for automated segmentation of viable and necrotic tumor on osteosarcoma whole slide images. One bottleneck for supervised learning is that large amounts of accurate annotations are required for training which is a time-consuming and expensive process. In this paper, we describe Deep Interactive Learning (DIaL) as an efficient labeling approach for training CNNs. After an initial labeling step is done, annotators only need to correct mislabeled regions from previous segmentation predictions to improve the CNN model until the satisfactory predictions are achieved. Our experiments show that our CNN model trained by only 7 hours of annotation using DIaL can successfully estimate ratios of necrosis within expected inter-observer variation rate for non-standardized manual surgical pathology task.
Deep Multi-Magnification Networks for Multi-Class Breast Cancer Image Segmentation
Oct 29, 2019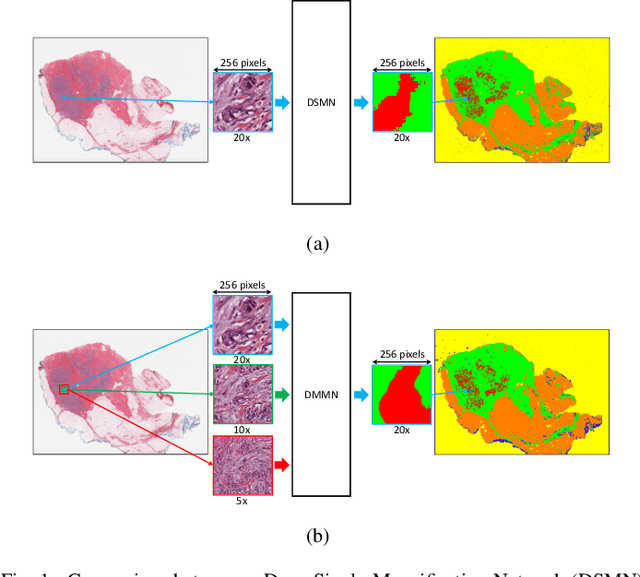
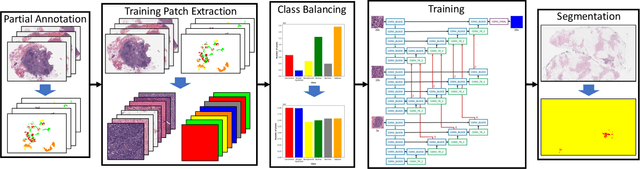
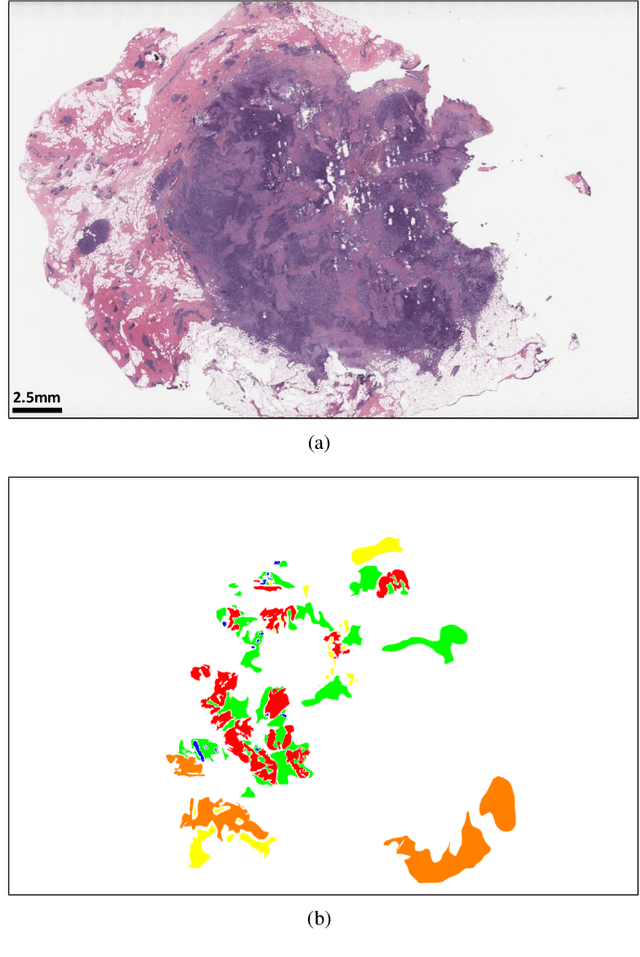
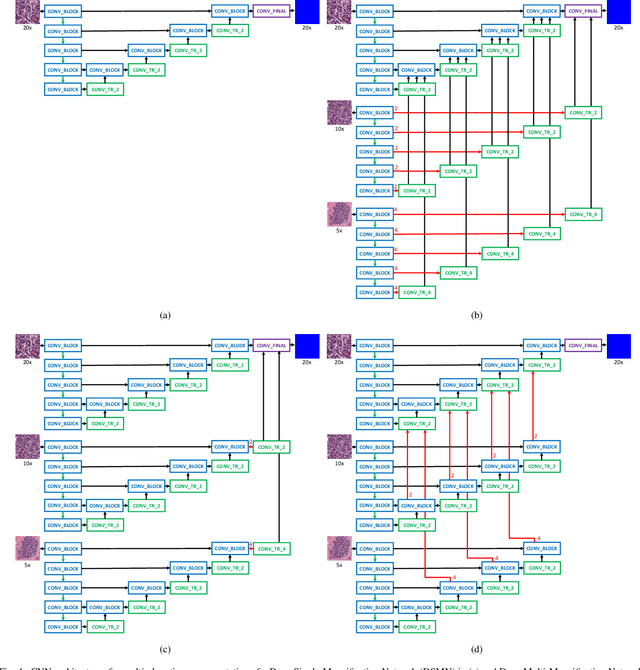
Abstract:Breast carcinoma is one of the most common cancers for women in the United States. Pathologic analysis of surgical excision specimens for breast carcinoma is important to evaluate the completeness of surgical excision and has implications for future treatment. This analysis is performed manually by pathologists reviewing histologic slides prepared from formalin-fixed tissue. Digital pathology has provided means to digitize the glass slides and generate whole slide images. Computational pathology enables whole slide images to be automatically analyzed to assist pathologists, especially with the advancement of deep learning. The whole slide images generally contain giga-pixels of data, so it is impractical to process the images at the whole-slide-level. Most of the current deep learning techniques process the images at the patch-level, but they may produce poor results by looking at individual patches with a narrow field-of-view at a single magnification. In this paper, we present Deep Multi-Magnification Networks (DMMNs) to resemble how pathologists analyze histologic slides using microscopes. Our multi-class tissue segmentation architecture processes a set of patches from multiple magnifications to make more accurate predictions. For our supervised training, we use partial annotations to reduce the burden of annotators. Our segmentation architecture with multi-encoder, multi-decoder, and multi-concatenation outperforms other segmentation architectures on breast datasets and can be used to facilitate pathologists' assessments of breast cancer.
Towards Unsupervised Cancer Subtyping: Predicting Prognosis Using A Histologic Visual Dictionary
Mar 12, 2019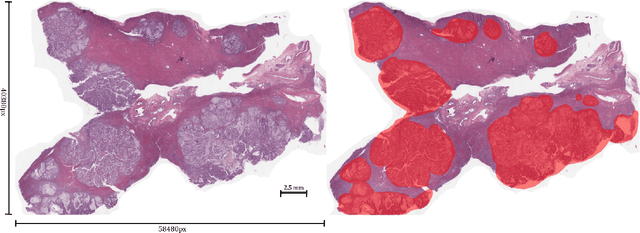
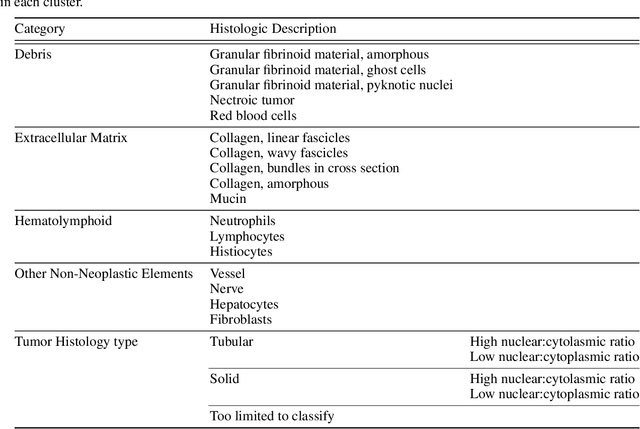
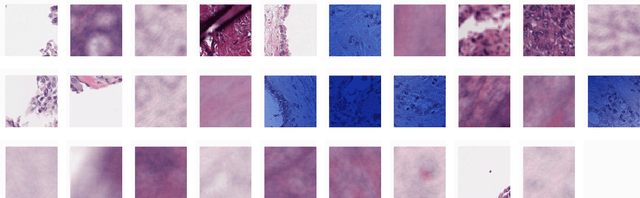
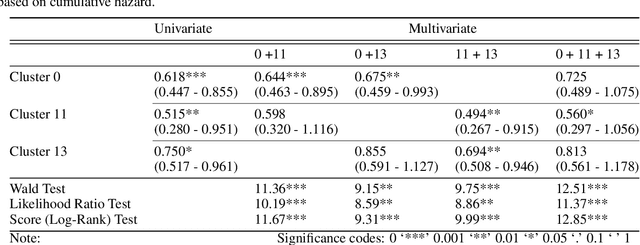
Abstract:Unlike common cancers, such as those of the prostate and breast, tumor grading in rare cancers is difficult and largely undefined because of small sample sizes, the sheer volume of time needed to undertake on such a task, and the inherent difficulty of extracting human-observed patterns. One of the most challenging examples is intrahepatic cholangiocarcinoma (ICC), a primary liver cancer arising from the biliary system, for which there is well-recognized tumor heterogeneity and no grading paradigm or prognostic biomarkers. In this paper, we propose a new unsupervised deep convolutional autoencoder-based clustering model that groups together cellular and structural morphologies of tumor in 246 ICC digitized whole slides, based on visual similarity. From this visual dictionary of histologic patterns, we use the clusters as covariates to train Cox-proportional hazard survival models. In univariate analysis, three clusters were significantly associated with recurrence-free survival. Combinations of these clusters were significant in multivariate analysis. In a multivariate analysis of all clusters, five showed significance to recurrence-free survival, however the overall model was not measured to be significant. Finally, a pathologist assigned clinical terminology to the significant clusters in the visual dictionary and found evidence supporting the hypothesis that collagen-enriched fibrosis plays a role in disease severity. These results offer insight into the future of cancer subtyping and show that computational pathology can contribute to disease prognostication, especially in rare cancers.
Terabyte-scale Deep Multiple Instance Learning for Classification and Localization in Pathology
Sep 27, 2018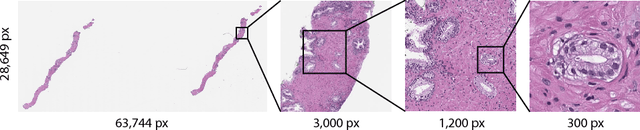
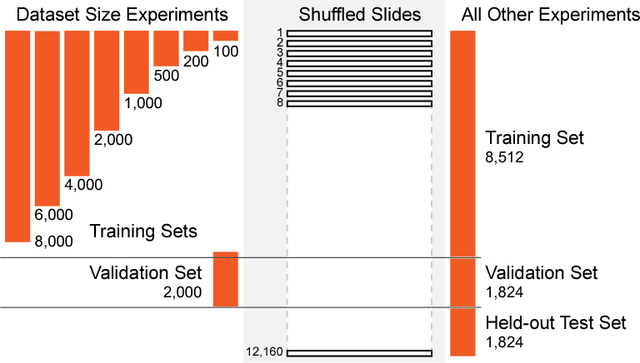
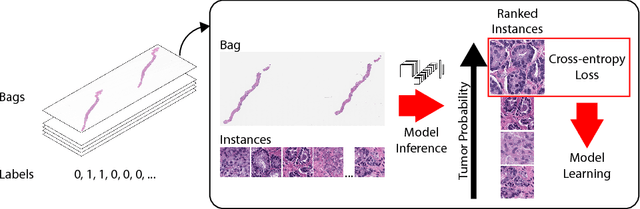
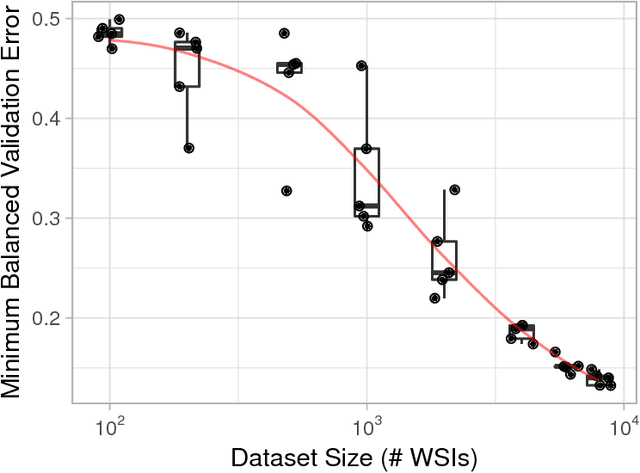
Abstract:In the field of computational pathology, the use of decision support systems powered by state-of-the-art deep learning solutions has been hampered by the lack of large labeled datasets. Until recently, studies relied on datasets in the order of few hundreds of slides which are not enough to train a model that can work at scale in the clinic. Here, we have gathered a dataset consisting of 12,160 slides, two orders of magnitude larger than previous datasets in pathology and equivalent to 25 times the pixel count of the entire ImageNet dataset. Given the size of our dataset it is possible for us to train a deep learning model under the Multiple Instance Learning (MIL) assumption where only the overall slide diagnosis is necessary for training, avoiding all the expensive pixel-wise annotations that are usually part of supervised learning approaches. We test our framework on a complex task, that of prostate cancer diagnosis on needle biopsies. We performed a thorough evaluation of the performance of our MIL pipeline under several conditions achieving an AUC of 0.98 on a held-out test set of 1,824 slides. These results open the way for training accurate diagnosis prediction models at scale, laying the foundation for decision support system deployment in the clinic.
DeepPET: A deep encoder-decoder network for directly solving the PET reconstruction inverse problem
Sep 25, 2018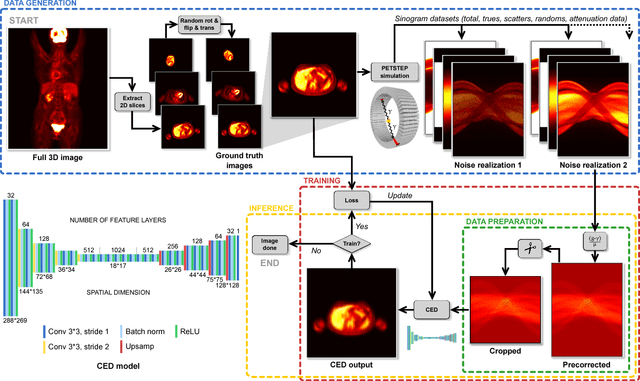
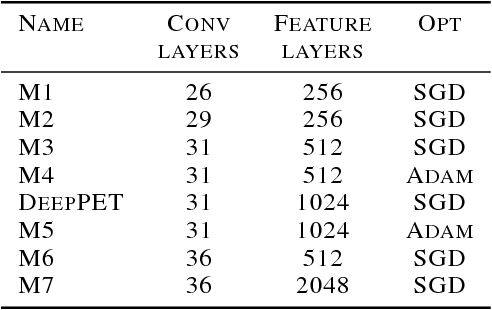
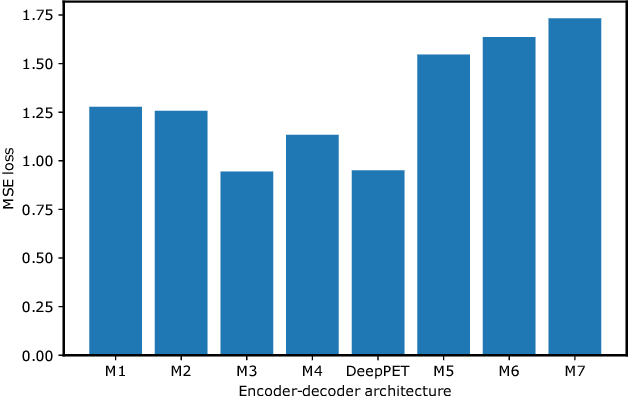
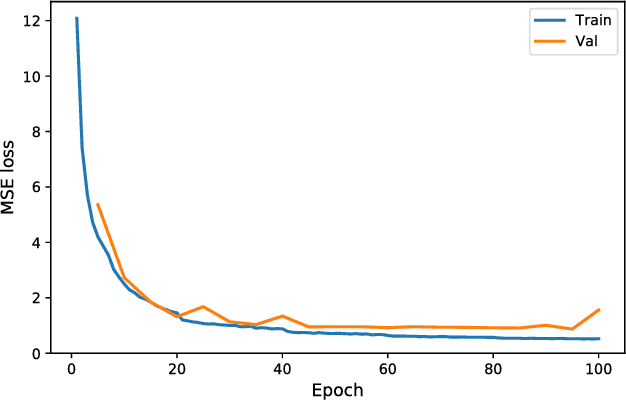
Abstract:Positron emission tomography (PET) is a cornerstone of modern radiology. The ability to detect cancer and metastases in whole body scans fundamentally changed cancer diagnosis and treatment. One of the main bottlenecks in the clinical application is the time it takes to reconstruct the anatomical image from the deluge of data in PET imaging. State-of-the art methods based on expectation maximization can take hours for a single patient and depend on manual fine-tuning. This results not only in financial burden for hospitals but more importantly leads to less efficient patient handling, evaluation, and ultimately diagnosis and treatment for patients. To overcome this problem we present a novel PET image reconstruction technique based on a deep convolutional encoder-decoder network, that takes PET sinogram data as input and directly outputs full PET images. Using realistic simulated data, we demonstrate that our network is able to reconstruct images >100 times faster, and with comparable image quality (in terms of root mean squared error) relative to conventional iterative reconstruction techniques.
Mitochondria-based Renal Cell Carcinoma Subtyping: Learning from Deep vs. Flat Feature Representations
Aug 02, 2016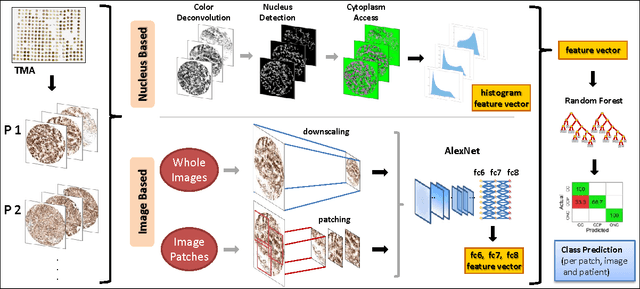
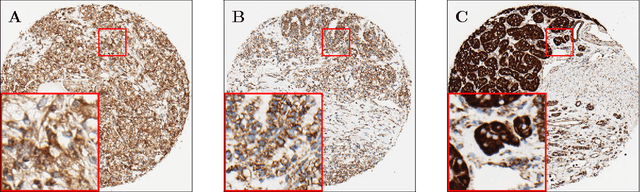
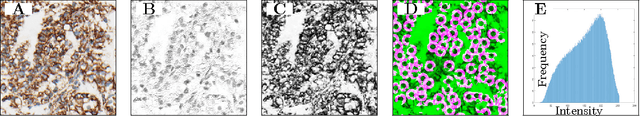
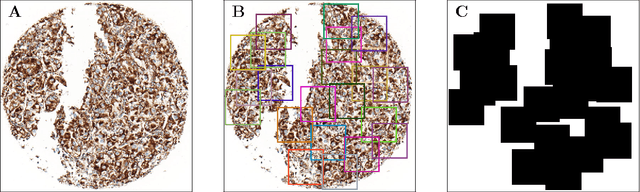
Abstract:Accurate subtyping of renal cell carcinoma (RCC) is of crucial importance for understanding disease progression and for making informed treatment decisions. New discoveries of significant alterations to mitochondria between subtypes make immunohistochemical (IHC) staining based image classification an imperative. Until now, accurate quantification and subtyping was made impossible by huge IHC variations, the absence of cell membrane staining for cytoplasm segmentation as well as the complete lack of systems for robust and reproducible image based classification. In this paper we present a comprehensive classification framework to overcome these challenges for tissue microarrays (TMA) of RCCs. We compare and evaluate models based on domain specific hand-crafted "flat"-features versus "deep" feature representations from various layers of a pre-trained convolutional neural network (CNN). The best model reaches a cross-validation accuracy of 89%, which demonstrates for the first time, that robust mitochondria-based subtyping of renal cancer is feasible
Computational Pathology: Challenges and Promises for Tissue Analysis
Dec 31, 2015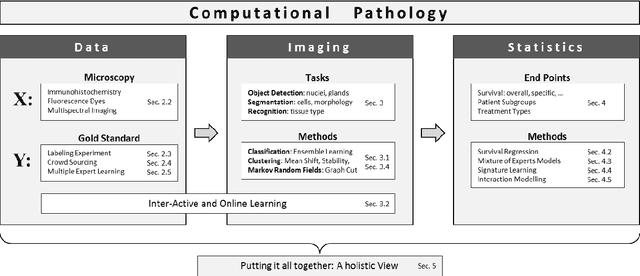
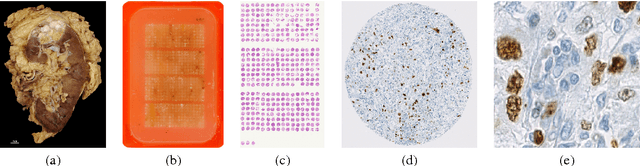
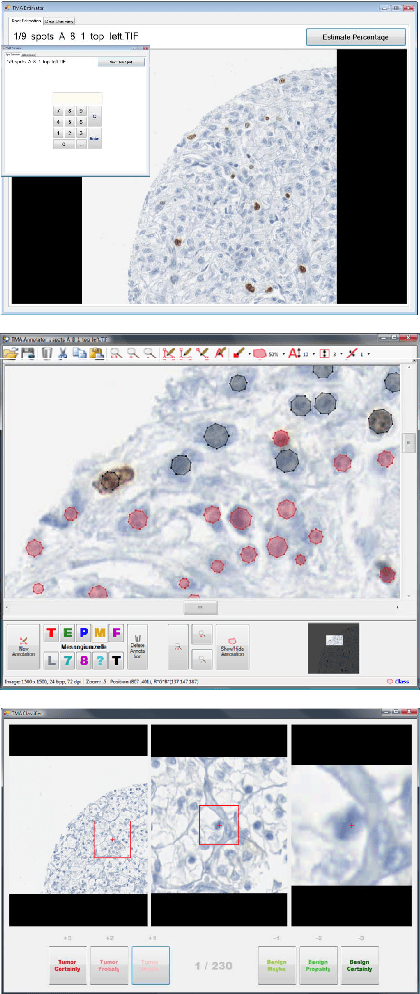
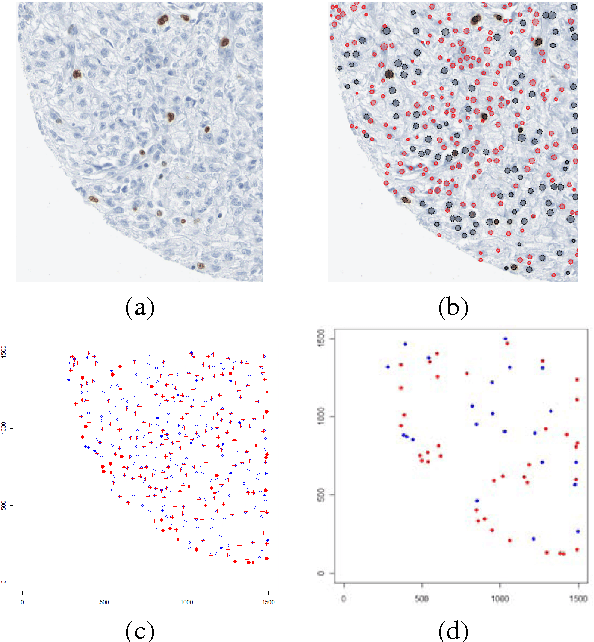
Abstract:The histological assessment of human tissue has emerged as the key challenge for detection and treatment of cancer. A plethora of different data sources ranging from tissue microarray data to gene expression, proteomics or metabolomics data provide a detailed overview of the health status of a patient. Medical doctors need to assess these information sources and they rely on data driven automatic analysis tools. Methods for classification, grouping and segmentation of heterogeneous data sources as well as regression of noisy dependencies and estimation of survival probabilities enter the processing workflow of a pathology diagnosis system at various stages. This paper reports on state-of-the-art of the design and effectiveness of computational pathology workflows and it discusses future research directions in this emergent field of medical informatics and diagnostic machine learning.
Early Recognition of Human Activities from First-Person Videos Using Onset Representations
Jul 06, 2015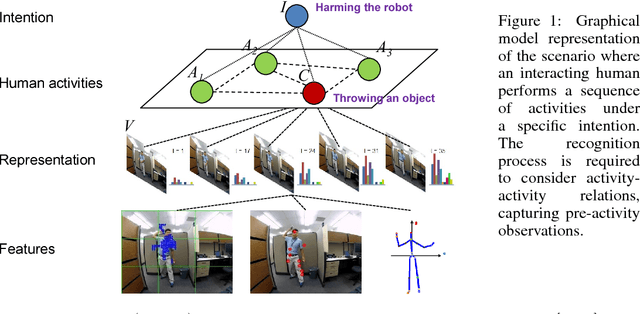
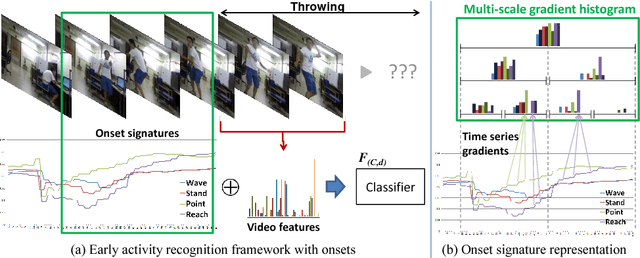

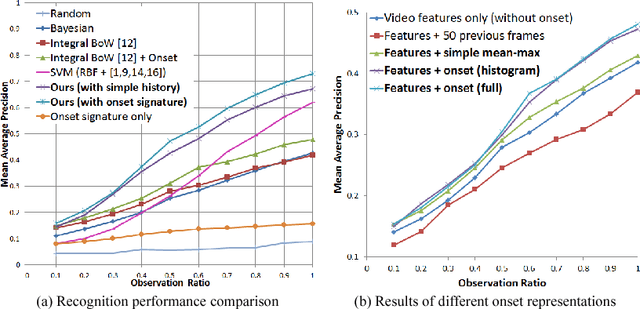
Abstract:In this paper, we propose a methodology for early recognition of human activities from videos taken with a first-person viewpoint. Early recognition, which is also known as activity prediction, is an ability to infer an ongoing activity at its early stage. We present an algorithm to perform recognition of activities targeted at the camera from streaming videos, making the system to predict intended activities of the interacting person and avoid harmful events before they actually happen. We introduce the novel concept of 'onset' that efficiently summarizes pre-activity observations, and design an approach to consider event history in addition to ongoing video observation for early first-person recognition of activities. We propose to represent onset using cascade histograms of time series gradients, and we describe a novel algorithmic setup to take advantage of onset for early recognition of activities. The experimental results clearly illustrate that the proposed concept of onset enables better/earlier recognition of human activities from first-person videos.
Boosting Convolutional Features for Robust Object Proposals
Mar 21, 2015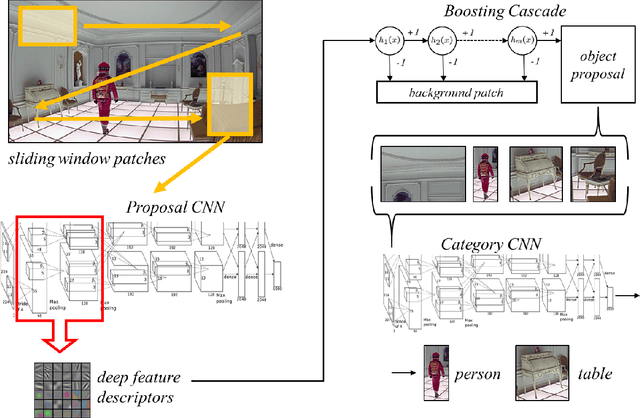
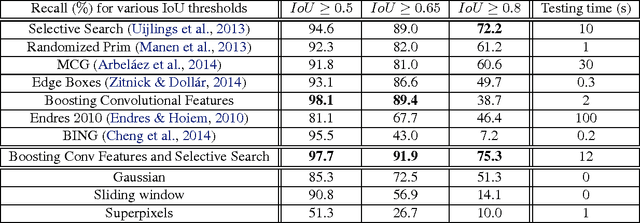
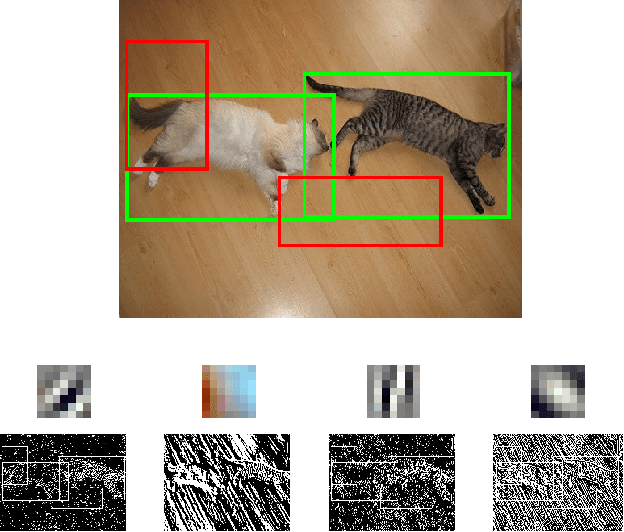
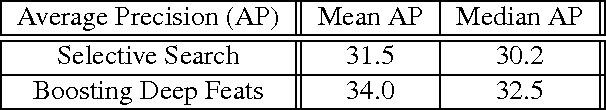
Abstract:Deep Convolutional Neural Networks (CNNs) have demonstrated excellent performance in image classification, but still show room for improvement in object-detection tasks with many categories, in particular for cluttered scenes and occlusion. Modern detection algorithms like Regions with CNNs (Girshick et al., 2014) rely on Selective Search (Uijlings et al., 2013) to propose regions which with high probability represent objects, where in turn CNNs are deployed for classification. Selective Search represents a family of sophisticated algorithms that are engineered with multiple segmentation, appearance and saliency cues, typically coming with a significant run-time overhead. Furthermore, (Hosang et al., 2014) have shown that most methods suffer from low reproducibility due to unstable superpixels, even for slight image perturbations. Although CNNs are subsequently used for classification in top-performing object-detection pipelines, current proposal methods are agnostic to how these models parse objects and their rich learned representations. As a result they may propose regions which may not resemble high-level objects or totally miss some of them. To overcome these drawbacks we propose a boosting approach which directly takes advantage of hierarchical CNN features for detecting regions of interest fast. We demonstrate its performance on ImageNet 2013 detection benchmark and compare it with state-of-the-art methods.
Feature Selection Strategies for Classifying High Dimensional Astronomical Data Sets
Oct 08, 2013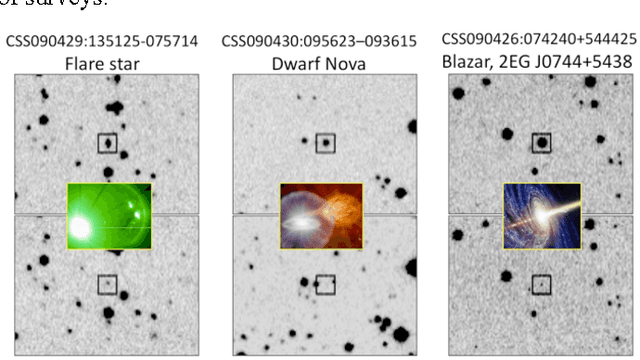
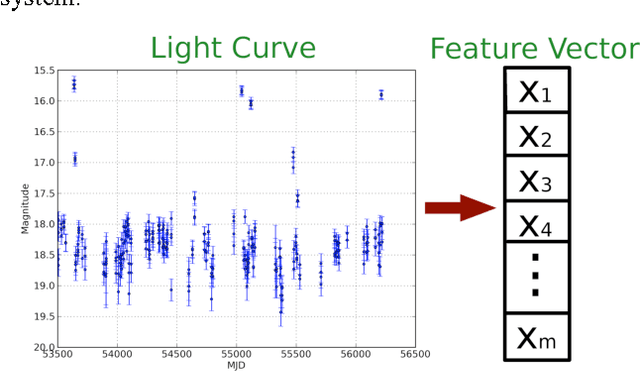
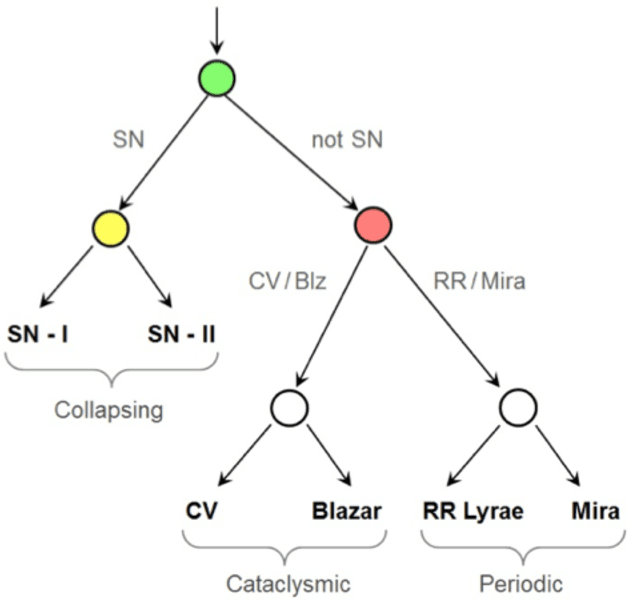
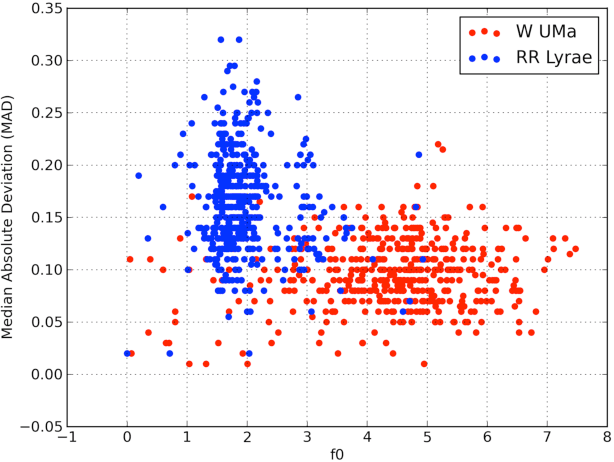
Abstract:The amount of collected data in many scientific fields is increasing, all of them requiring a common task: extract knowledge from massive, multi parametric data sets, as rapidly and efficiently possible. This is especially true in astronomy where synoptic sky surveys are enabling new research frontiers in the time domain astronomy and posing several new object classification challenges in multi dimensional spaces; given the high number of parameters available for each object, feature selection is quickly becoming a crucial task in analyzing astronomical data sets. Using data sets extracted from the ongoing Catalina Real-Time Transient Surveys (CRTS) and the Kepler Mission we illustrate a variety of feature selection strategies used to identify the subsets that give the most information and the results achieved applying these techniques to three major astronomical problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge