Gabriele Campanella
Predict Patient Self-reported Race from Skin Histological Images
Jul 30, 2025Abstract:Artificial Intelligence (AI) has demonstrated success in computational pathology (CPath) for disease detection, biomarker classification, and prognosis prediction. However, its potential to learn unintended demographic biases, particularly those related to social determinants of health, remains understudied. This study investigates whether deep learning models can predict self-reported race from digitized dermatopathology slides and identifies potential morphological shortcuts. Using a multisite dataset with a racially diverse population, we apply an attention-based mechanism to uncover race-associated morphological features. After evaluating three dataset curation strategies to control for confounding factors, the final experiment showed that White and Black demographic groups retained high prediction performance (AUC: 0.799, 0.762), while overall performance dropped to 0.663. Attention analysis revealed the epidermis as a key predictive feature, with significant performance declines when these regions were removed. These findings highlight the need for careful data curation and bias mitigation to ensure equitable AI deployment in pathology. Code available at: https://github.com/sinai-computational-pathology/CPath_SAIF.
Single GPU Task Adaptation of Pathology Foundation Models for Whole Slide Image Analysis
Jun 05, 2025
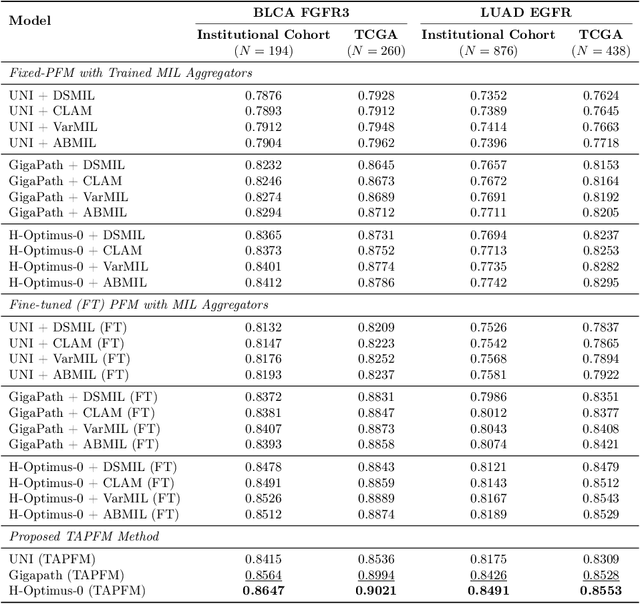
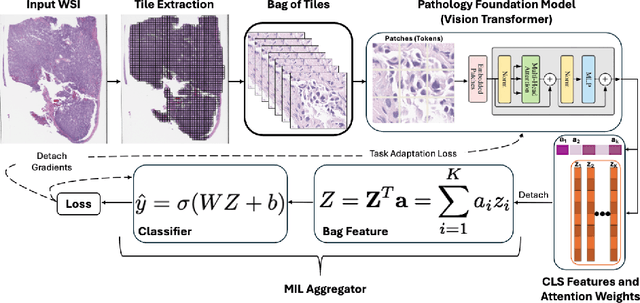
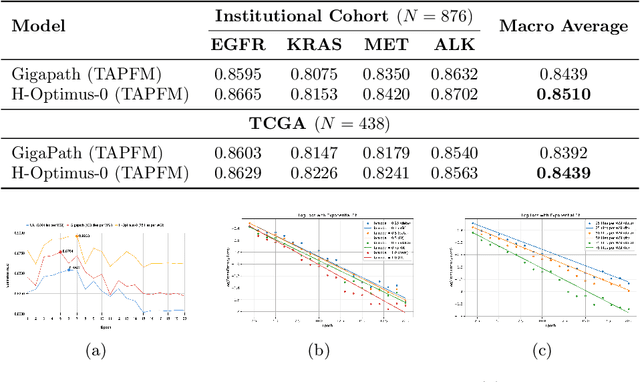
Abstract:Pathology foundation models (PFMs) have emerged as powerful tools for analyzing whole slide images (WSIs). However, adapting these pretrained PFMs for specific clinical tasks presents considerable challenges, primarily due to the availability of only weak (WSI-level) labels for gigapixel images, necessitating multiple instance learning (MIL) paradigm for effective WSI analysis. This paper proposes a novel approach for single-GPU \textbf{T}ask \textbf{A}daptation of \textbf{PFM}s (TAPFM) that uses vision transformer (\vit) attention for MIL aggregation while optimizing both for feature representations and attention weights. The proposed approach maintains separate computational graphs for MIL aggregator and the PFM to create stable training dynamics that align with downstream task objectives during end-to-end adaptation. Evaluated on mutation prediction tasks for bladder cancer and lung adenocarcinoma across institutional and TCGA cohorts, TAPFM consistently outperforms conventional approaches, with H-Optimus-0 (TAPFM) outperforming the benchmarks. TAPFM effectively handles multi-label classification of actionable mutations as well. Thus, TAPFM makes adaptation of powerful pre-trained PFMs practical on standard hardware for various clinical applications.
Revisiting Automatic Data Curation for Vision Foundation Models in Digital Pathology
Mar 24, 2025Abstract:Vision foundation models (FMs) are accelerating the development of digital pathology algorithms and transforming biomedical research. These models learn, in a self-supervised manner, to represent histological features in highly heterogeneous tiles extracted from whole-slide images (WSIs) of real-world patient samples. The performance of these FMs is significantly influenced by the size, diversity, and balance of the pre-training data. However, data selection has been primarily guided by expert knowledge at the WSI level, focusing on factors such as disease classification and tissue types, while largely overlooking the granular details available at the tile level. In this paper, we investigate the potential of unsupervised automatic data curation at the tile-level, taking into account 350 million tiles. Specifically, we apply hierarchical clustering trees to pre-extracted tile embeddings, allowing us to sample balanced datasets uniformly across the embedding space of the pretrained FM. We further identify these datasets are subject to a trade-off between size and balance, potentially compromising the quality of representations learned by FMs, and propose tailored batch sampling strategies to mitigate this effect. We demonstrate the effectiveness of our method through improved performance on a diverse range of clinically relevant downstream tasks.
A Clinical Benchmark of Public Self-Supervised Pathology Foundation Models
Jul 11, 2024



Abstract:The use of self-supervised learning (SSL) to train pathology foundation models has increased substantially in the past few years. Notably, several models trained on large quantities of clinical data have been made publicly available in recent months. This will significantly enhance scientific research in computational pathology and help bridge the gap between research and clinical deployment. With the increase in availability of public foundation models of different sizes, trained using different algorithms on different datasets, it becomes important to establish a benchmark to compare the performance of such models on a variety of clinically relevant tasks spanning multiple organs and diseases. In this work, we present a collection of pathology datasets comprising clinical slides associated with clinically relevant endpoints including cancer diagnoses and a variety of biomarkers generated during standard hospital operation from two medical centers. We leverage these datasets to systematically assess the performance of public pathology foundation models and provide insights into best practices for training new foundation models and selecting appropriate pretrained models.
Benchmarking Embedding Aggregation Methods in Computational Pathology: A Clinical Data Perspective
Jul 10, 2024


Abstract:Recent advances in artificial intelligence (AI), in particular self-supervised learning of foundation models (FMs), are revolutionizing medical imaging and computational pathology (CPath). A constant challenge in the analysis of digital Whole Slide Images (WSIs) is the problem of aggregating tens of thousands of tile-level image embeddings to a slide-level representation. Due to the prevalent use of datasets created for genomic research, such as TCGA, for method development, the performance of these techniques on diagnostic slides from clinical practice has been inadequately explored. This study conducts a thorough benchmarking analysis of ten slide-level aggregation techniques across nine clinically relevant tasks, including diagnostic assessment, biomarker classification, and outcome prediction. The results yield following key insights: (1) Embeddings derived from domain-specific (histological images) FMs outperform those from generic ImageNet-based models across aggregation methods. (2) Spatial-aware aggregators enhance the performance significantly when using ImageNet pre-trained models but not when using FMs. (3) No single model excels in all tasks and spatially-aware models do not show general superiority as it would be expected. These findings underscore the need for more adaptable and universally applicable aggregation techniques, guiding future research towards tools that better meet the evolving needs of clinical-AI in pathology. The code used in this work is available at \url{https://github.com/fuchs-lab-public/CPath_SABenchmark}.
Beyond Multiple Instance Learning: Full Resolution All-In-Memory End-To-End Pathology Slide Modeling
Mar 07, 2024



Abstract:Artificial Intelligence (AI) has great potential to improve health outcomes by training systems on vast digitized clinical datasets. Computational Pathology, with its massive amounts of microscopy image data and impact on diagnostics and biomarkers, is at the forefront of this development. Gigapixel pathology slides pose a unique challenge due to their enormous size and are usually divided into tens of thousands of smaller tiles for analysis. This results in a discontinuity in the machine learning process by separating the training of tile-level encoders from slide-level aggregators and the need to adopt weakly supervised learning strategies. Training models from entire pathology slides end-to-end has been largely unexplored due to its computational challenges. To overcome this problem, we propose a novel approach to jointly train both a tile encoder and a slide-aggregator fully in memory and end-to-end at high-resolution, bridging the gap between input and slide-level supervision. While more computationally expensive, detailed quantitative validation shows promise for large-scale pre-training of pathology foundation models.
Computational Pathology at Health System Scale -- Self-Supervised Foundation Models from Three Billion Images
Oct 10, 2023Abstract:Recent breakthroughs in self-supervised learning have enabled the use of large unlabeled datasets to train visual foundation models that can generalize to a variety of downstream tasks. While this training paradigm is well suited for the medical domain where annotations are scarce, large-scale pre-training in the medical domain, and in particular pathology, has not been extensively studied. Previous work in self-supervised learning in pathology has leveraged smaller datasets for both pre-training and evaluating downstream performance. The aim of this project is to train the largest academic foundation model and benchmark the most prominent self-supervised learning algorithms by pre-training and evaluating downstream performance on large clinical pathology datasets. We collected the largest pathology dataset to date, consisting of over 3 billion images from over 423 thousand microscopy slides. We compared pre-training of visual transformer models using the masked autoencoder (MAE) and DINO algorithms. We evaluated performance on six clinically relevant tasks from three anatomic sites and two institutions: breast cancer detection, inflammatory bowel disease detection, breast cancer estrogen receptor prediction, lung adenocarcinoma EGFR mutation prediction, and lung cancer immunotherapy response prediction. Our results demonstrate that pre-training on pathology data is beneficial for downstream performance compared to pre-training on natural images. Additionally, the DINO algorithm achieved better generalization performance across all tasks tested. The presented results signify a phase change in computational pathology research, paving the way into a new era of more performant models based on large-scale, parallel pre-training at the billion-image scale.
Deep conditional transformation models for survival analysis
Oct 20, 2022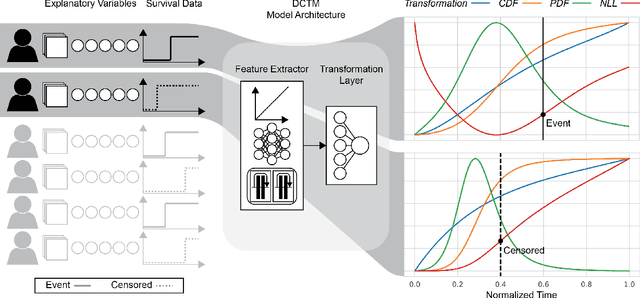
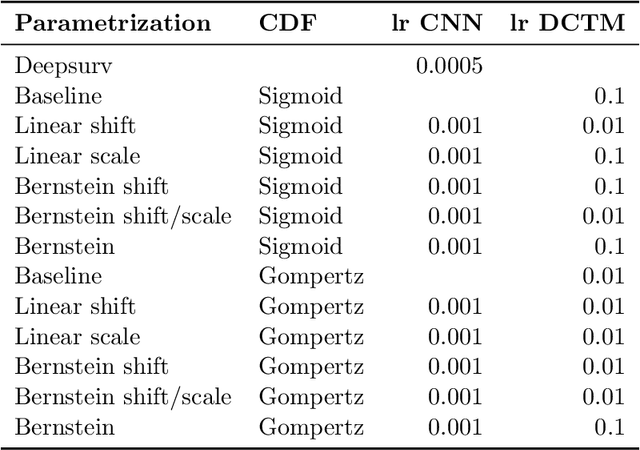
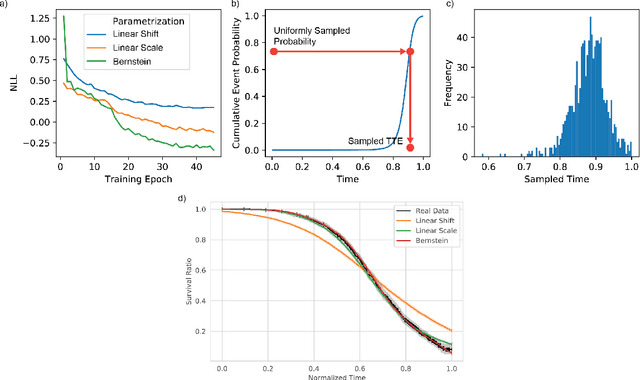
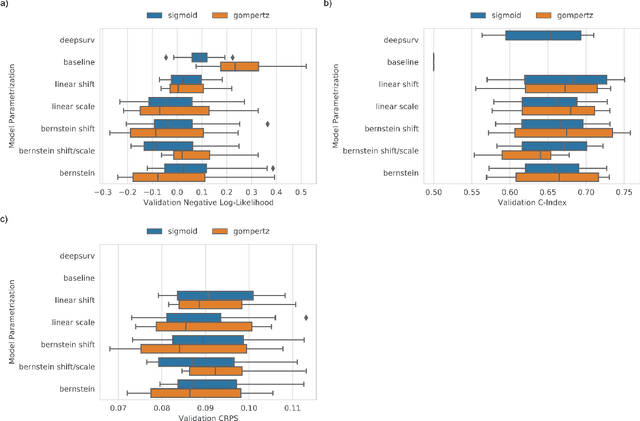
Abstract:An every increasing number of clinical trials features a time-to-event outcome and records non-tabular patient data, such as magnetic resonance imaging or text data in the form of electronic health records. Recently, several neural-network based solutions have been proposed, some of which are binary classifiers. Parametric, distribution-free approaches which make full use of survival time and censoring status have not received much attention. We present deep conditional transformation models (DCTMs) for survival outcomes as a unifying approach to parametric and semiparametric survival analysis. DCTMs allow the specification of non-linear and non-proportional hazards for both tabular and non-tabular data and extend to all types of censoring and truncation. On real and semi-synthetic data, we show that DCTMs compete with state-of-the-art DL approaches to survival analysis.
H&E-based Computational Biomarker Enables Universal EGFR Screening for Lung Adenocarcinoma
Jun 21, 2022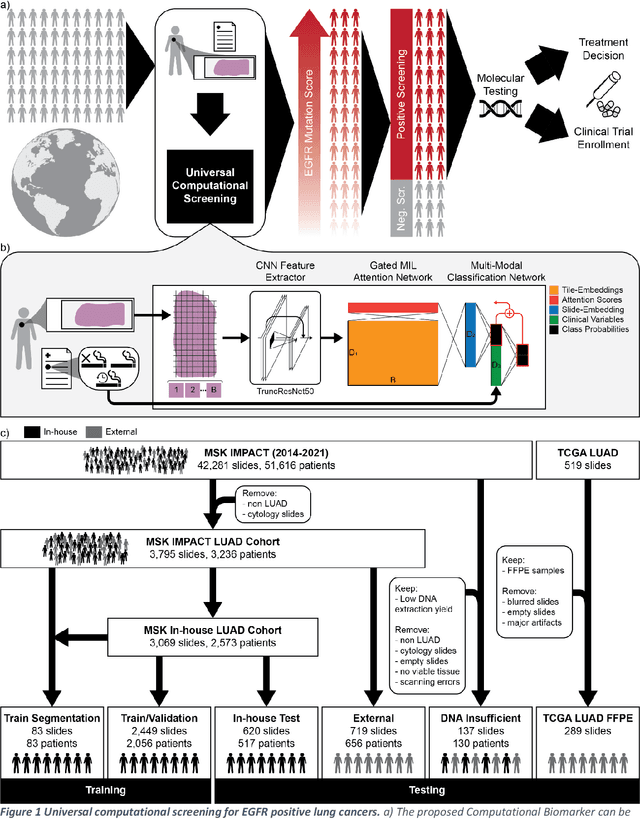
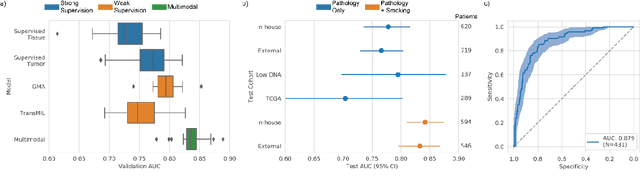
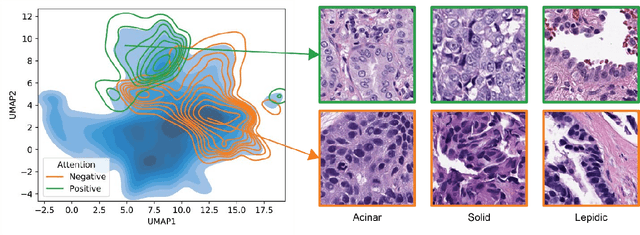
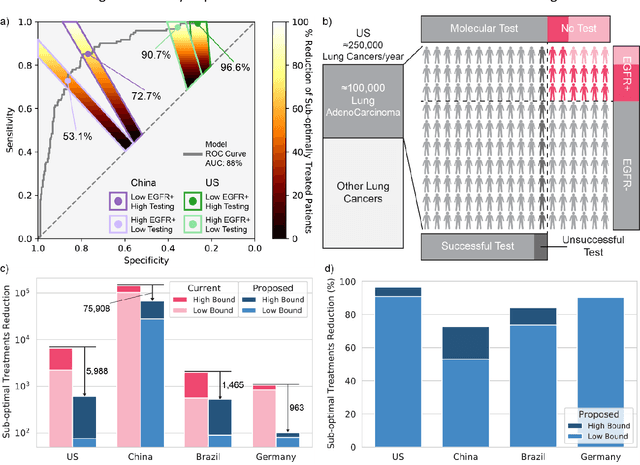
Abstract:Lung cancer is the leading cause of cancer death worldwide, with lung adenocarcinoma being the most prevalent form of lung cancer. EGFR positive lung adenocarcinomas have been shown to have high response rates to TKI therapy, underlying the essential nature of molecular testing for lung cancers. Despite current guidelines consider testing necessary, a large portion of patients are not routinely profiled, resulting in millions of people not receiving the optimal treatment for their lung cancer. Sequencing is the gold standard for molecular testing of EGFR mutations, but it can take several weeks for results to come back, which is not ideal in a time constrained scenario. The development of alternative screening tools capable of detecting EGFR mutations quickly and cheaply while preserving tissue for sequencing could help reduce the amount of sub-optimally treated patients. We propose a multi-modal approach which integrates pathology images and clinical variables to predict EGFR mutational status achieving an AUC of 84% on the largest clinical cohort to date. Such a computational model could be deployed at large at little additional cost. Its clinical application could reduce the number of patients who receive sub-optimal treatments by 53.1% in China, and up to 96.6% in the US.
Towards Unsupervised Cancer Subtyping: Predicting Prognosis Using A Histologic Visual Dictionary
Mar 12, 2019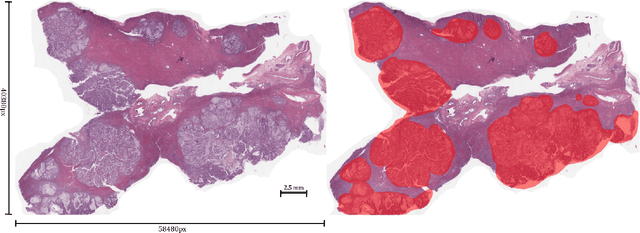
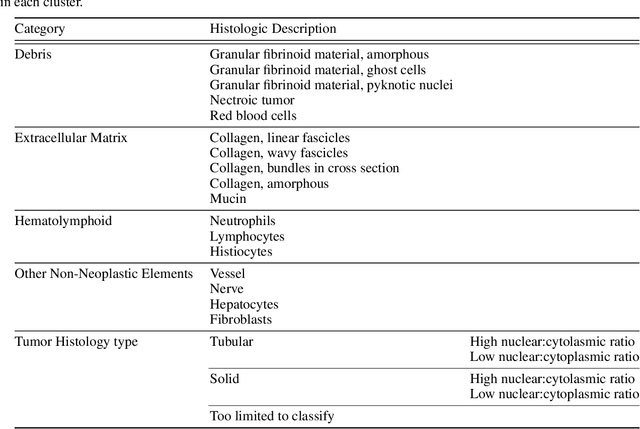
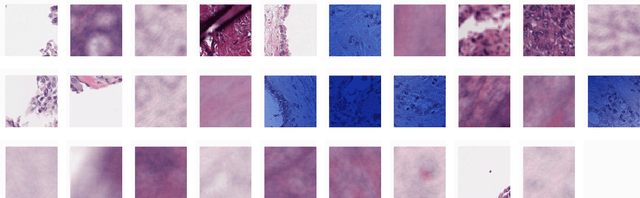
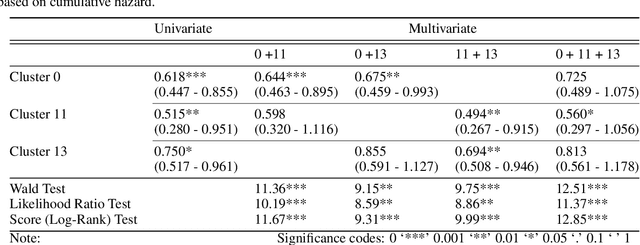
Abstract:Unlike common cancers, such as those of the prostate and breast, tumor grading in rare cancers is difficult and largely undefined because of small sample sizes, the sheer volume of time needed to undertake on such a task, and the inherent difficulty of extracting human-observed patterns. One of the most challenging examples is intrahepatic cholangiocarcinoma (ICC), a primary liver cancer arising from the biliary system, for which there is well-recognized tumor heterogeneity and no grading paradigm or prognostic biomarkers. In this paper, we propose a new unsupervised deep convolutional autoencoder-based clustering model that groups together cellular and structural morphologies of tumor in 246 ICC digitized whole slides, based on visual similarity. From this visual dictionary of histologic patterns, we use the clusters as covariates to train Cox-proportional hazard survival models. In univariate analysis, three clusters were significantly associated with recurrence-free survival. Combinations of these clusters were significant in multivariate analysis. In a multivariate analysis of all clusters, five showed significance to recurrence-free survival, however the overall model was not measured to be significant. Finally, a pathologist assigned clinical terminology to the significant clusters in the visual dictionary and found evidence supporting the hypothesis that collagen-enriched fibrosis plays a role in disease severity. These results offer insight into the future of cancer subtyping and show that computational pathology can contribute to disease prognostication, especially in rare cancers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge