Shuaipeng Zhang
DAG-AFL:Directed Acyclic Graph-based Asynchronous Federated Learning
Jul 28, 2025Abstract:Due to the distributed nature of federated learning (FL), the vulnerability of the global model and the need for coordination among many client devices pose significant challenges. As a promising decentralized, scalable and secure solution, blockchain-based FL methods have attracted widespread attention in recent years. However, traditional consensus mechanisms designed for Proof of Work (PoW) similar to blockchain incur substantial resource consumption and compromise the efficiency of FL, particularly when participating devices are wireless and resource-limited. To address asynchronous client participation and data heterogeneity in FL, while limiting the additional resource overhead introduced by blockchain, we propose the Directed Acyclic Graph-based Asynchronous Federated Learning (DAG-AFL) framework. We develop a tip selection algorithm that considers temporal freshness, node reachability and model accuracy, with a DAG-based trusted verification strategy. Extensive experiments on 3 benchmarking datasets against eight state-of-the-art approaches demonstrate that DAG-AFL significantly improves training efficiency and model accuracy by 22.7% and 6.5% on average, respectively.
MolMiner: You only look once for chemical structure recognition
May 23, 2022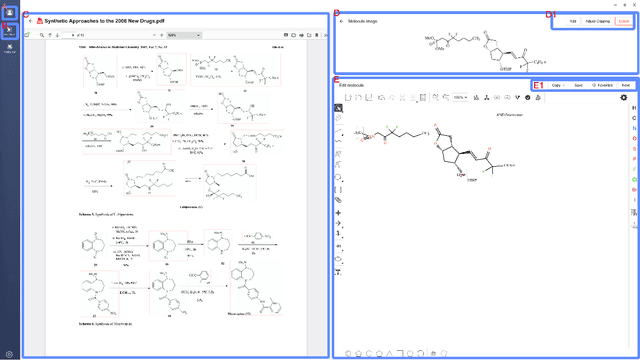
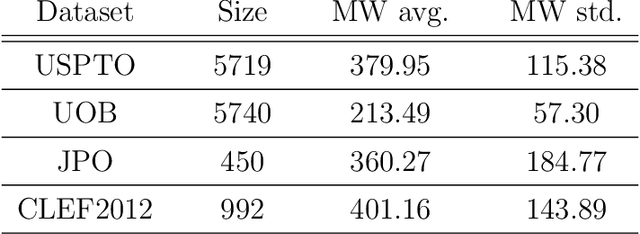
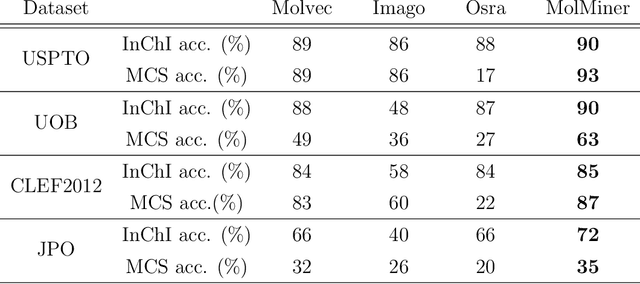
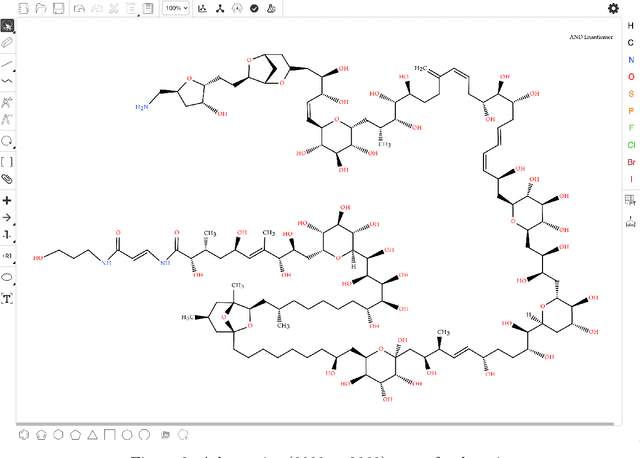
Abstract:Molecular structures are always depicted as 2D printed form in scientific documents like journal papers and patents. However, these 2D depictions are not machine-readable. Due to a backlog of decades and an increasing amount of these printed literature, there is a high demand for the translation of printed depictions into machine-readable formats, which is known as Optical Chemical Structure Recognition (OCSR). Most OCSR systems developed over the last three decades follow a rule-based approach where the key step of vectorization of the depiction is based on the interpretation of vectors and nodes as bonds and atoms. Here, we present a practical software MolMiner, which is primarily built up using deep neural networks originally developed for semantic segmentation and object detection to recognize atom and bond elements from documents. These recognized elements can be easily connected as a molecular graph with distance-based construction algorithm. We carefully evaluate our software on four benchmark datasets with the state-of-the-art performance. Various real application scenarios are also tested, yielding satisfactory outcomes. The free download links of Mac and Windows versions are available: Mac: https://molminer-cdn.iipharma.cn/pharma-mind/artifact/latest/mac/PharmaMind-mac-latest-setup.dmg and Windows: https://molminer-cdn.iipharma.cn/pharma-mind/artifact/latest/win/PharmaMind-win-latest-setup.exe
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge