Jianhang Zhang
Bridging Vision and Language for Robust Context-Aware Surgical Point Tracking: The VL-SurgPT Dataset and Benchmark
Nov 15, 2025Abstract:Accurate point tracking in surgical environments remains challenging due to complex visual conditions, including smoke occlusion, specular reflections, and tissue deformation. While existing surgical tracking datasets provide coordinate information, they lack the semantic context necessary to understand tracking failure mechanisms. We introduce VL-SurgPT, the first large-scale multimodal dataset that bridges visual tracking with textual descriptions of point status in surgical scenes. The dataset comprises 908 in vivo video clips, including 754 for tissue tracking (17,171 annotated points across five challenging scenarios) and 154 for instrument tracking (covering seven instrument types with detailed keypoint annotations). We establish comprehensive benchmarks using eight state-of-the-art tracking methods and propose TG-SurgPT, a text-guided tracking approach that leverages semantic descriptions to improve robustness in visually challenging conditions. Experimental results demonstrate that incorporating point status information significantly improves tracking accuracy and reliability, particularly in adverse visual scenarios where conventional vision-only methods struggle. By bridging visual and linguistic modalities, VL-SurgPT enables the development of context-aware tracking systems crucial for advancing computer-assisted surgery applications that can maintain performance even under challenging intraoperative conditions.
MMME: A Spontaneous Multi-Modal Micro-Expression Dataset Enabling Visual-Physiological Fusion
Jun 12, 2025Abstract:Micro-expressions (MEs) are subtle, fleeting nonverbal cues that reveal an individual's genuine emotional state. Their analysis has attracted considerable interest due to its promising applications in fields such as healthcare, criminal investigation, and human-computer interaction. However, existing ME research is limited to single visual modality, overlooking the rich emotional information conveyed by other physiological modalities, resulting in ME recognition and spotting performance far below practical application needs. Therefore, exploring the cross-modal association mechanism between ME visual features and physiological signals (PS), and developing a multimodal fusion framework, represents a pivotal step toward advancing ME analysis. This study introduces a novel ME dataset, MMME, which, for the first time, enables synchronized collection of facial action signals (MEs), central nervous system signals (EEG), and peripheral PS (PPG, RSP, SKT, EDA, and ECG). By overcoming the constraints of existing ME corpora, MMME comprises 634 MEs, 2,841 macro-expressions (MaEs), and 2,890 trials of synchronized multimodal PS, establishing a robust foundation for investigating ME neural mechanisms and conducting multimodal fusion-based analyses. Extensive experiments validate the dataset's reliability and provide benchmarks for ME analysis, demonstrating that integrating MEs with PS significantly enhances recognition and spotting performance. To the best of our knowledge, MMME is the most comprehensive ME dataset to date in terms of modality diversity. It provides critical data support for exploring the neural mechanisms of MEs and uncovering the visual-physiological synergistic effects, driving a paradigm shift in ME research from single-modality visual analysis to multimodal fusion. The dataset will be publicly available upon acceptance of this paper.
MolMiner: You only look once for chemical structure recognition
May 23, 2022
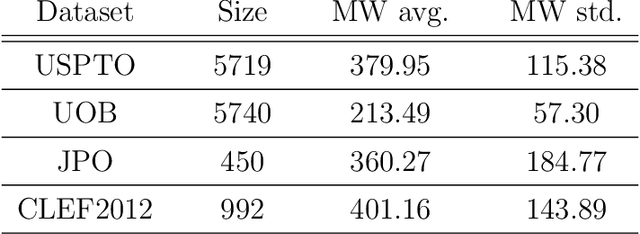
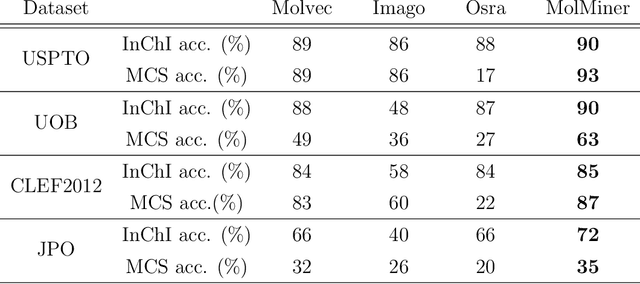
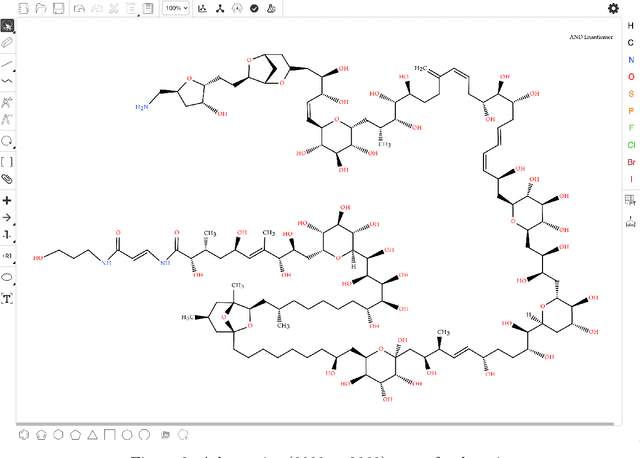
Abstract:Molecular structures are always depicted as 2D printed form in scientific documents like journal papers and patents. However, these 2D depictions are not machine-readable. Due to a backlog of decades and an increasing amount of these printed literature, there is a high demand for the translation of printed depictions into machine-readable formats, which is known as Optical Chemical Structure Recognition (OCSR). Most OCSR systems developed over the last three decades follow a rule-based approach where the key step of vectorization of the depiction is based on the interpretation of vectors and nodes as bonds and atoms. Here, we present a practical software MolMiner, which is primarily built up using deep neural networks originally developed for semantic segmentation and object detection to recognize atom and bond elements from documents. These recognized elements can be easily connected as a molecular graph with distance-based construction algorithm. We carefully evaluate our software on four benchmark datasets with the state-of-the-art performance. Various real application scenarios are also tested, yielding satisfactory outcomes. The free download links of Mac and Windows versions are available: Mac: https://molminer-cdn.iipharma.cn/pharma-mind/artifact/latest/mac/PharmaMind-mac-latest-setup.dmg and Windows: https://molminer-cdn.iipharma.cn/pharma-mind/artifact/latest/win/PharmaMind-win-latest-setup.exe
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge