MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023Jianning Li, Antonio Pepe, Christina Gsaxner, Gijs Luijten, Yuan Jin, Narmada Ambigapathy, Enrico Nasca, Naida Solak, Gian Marco Melito, Viet Duc Vu, Afaque R. Memon, Xiaojun Chen, Jan Stefan Kirschke, Ezequiel de la Rosa, Patrick Ferdinand Christ, Hongwei Bran Li, David G. Ellis, Michele R. Aizenberg, Sergios Gatidis, Thomas Küstner, Nadya Shusharina, Nicholas Heller, Vincent Andrearczyk, Adrien Depeursinge, Mathieu Hatt, Anjany Sekuboyina, Maximilian Löffler, Hans Liebl, Reuben Dorent, Tom Vercauteren, Jonathan Shapey, Aaron Kujawa, Stefan Cornelissen, Patrick Langenhuizen, Achraf Ben-Hamadou, Ahmed Rekik, Sergi Pujades, Edmond Boyer, Federico Bolelli, Costantino Grana, Luca Lumetti, Hamidreza Salehi, Jun Ma, Yao Zhang, Ramtin Gharleghi, Susann Beier, Arcot Sowmya, Eduardo A. Garza-Villarreal, Thania Balducci, Diego Angeles-Valdez, Roberto Souza, Leticia Rittner, Richard Frayne, Yuanfeng Ji, Soumick Chatterjee, Florian Dubost, Stefanie Schreiber, Hendrik Mattern, Oliver Speck, Daniel Haehn, Christoph John, Andreas Nürnberger, João Pedrosa, Carlos Ferreira, Guilherme Aresta, António Cunha, Aurélio Campilho, Yannick Suter, Jose Garcia, Alain Lalande, Emmanuel Audenaert, Claudia Krebs, Timo Van Leeuwen, Evie Vereecke, Rainer Röhrig, Frank Hölzle, Vahid Badeli, Kathrin Krieger, Matthias Gunzer, Jianxu Chen, Amin Dada, Miriam Balzer, Jana Fragemann, Frederic Jonske, Moritz Rempe, Stanislav Malorodov, Fin H. Bahnsen, Constantin Seibold, Alexander Jaus, Ana Sofia Santos, Mariana Lindo, André Ferreira, Victor Alves, Michael Kamp, Amr Abourayya, Felix Nensa, Fabian Hörst, Alexander Brehmer, Lukas Heine, Lars E. Podleska, Matthias A. Fink, Julius Keyl, Konstantinos Tserpes, Moon-Sung Kim, Shireen Elhabian, Hans Lamecker, Dženan Zukić, Beatriz Paniagua, Christian Wachinger, Martin Urschler, Luc Duong, Jakob Wasserthal, Peter F. Hoyer, Oliver Basu, Thomas Maal, Max J. H. Witjes, Ti-chiun Chang, Seyed-Ahmad Ahmadi, Ping Luo, Bjoern Menze, Mauricio Reyes, Christos Davatzikos, Behrus Puladi, Jens Kleesiek, Jan Egger
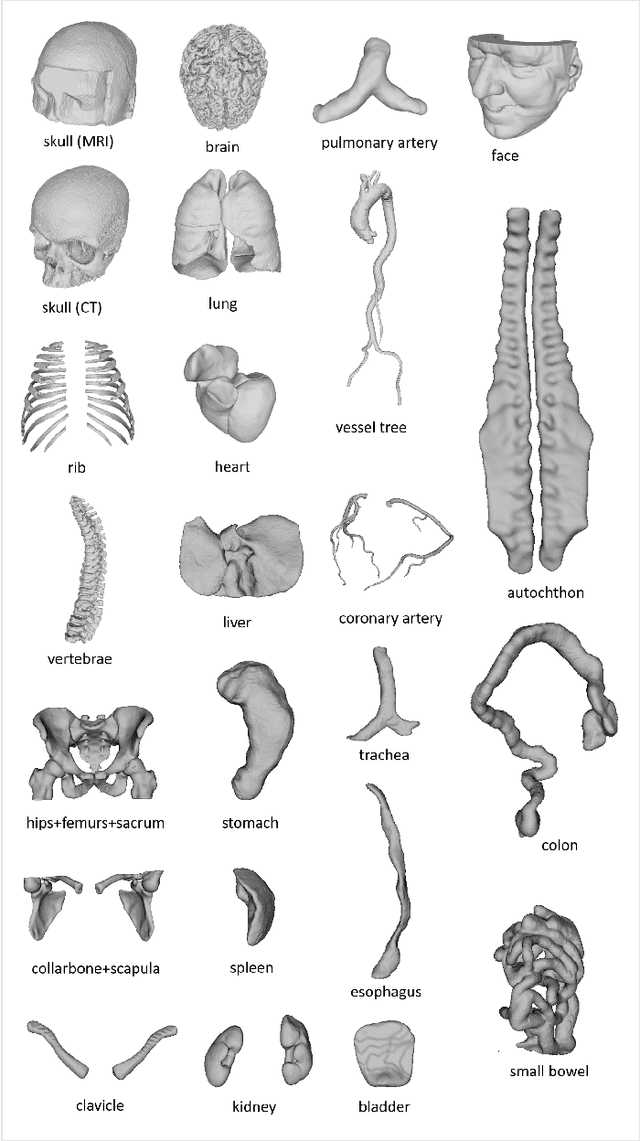

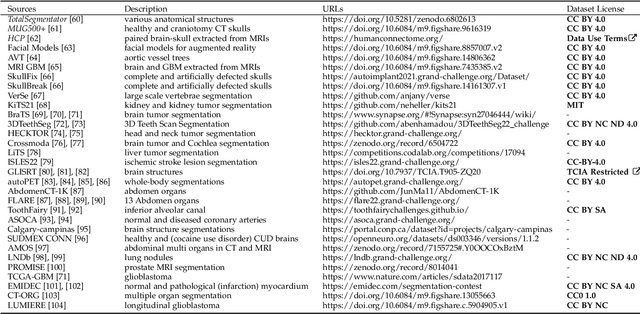
We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
DIAMANT: Dual Image-Attention Map Encoders For Medical Image Segmentation
Apr 28, 2023Yousef Yeganeh, Azade Farshad, Peter Weinberger, Seyed-Ahmad Ahmadi, Ehsan Adeli, Nassir Navab
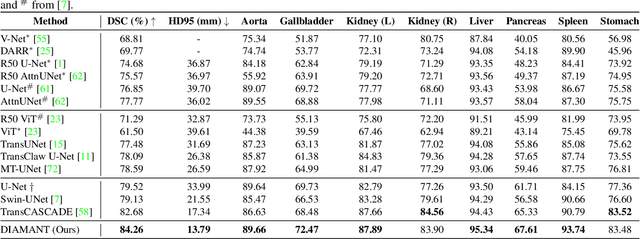
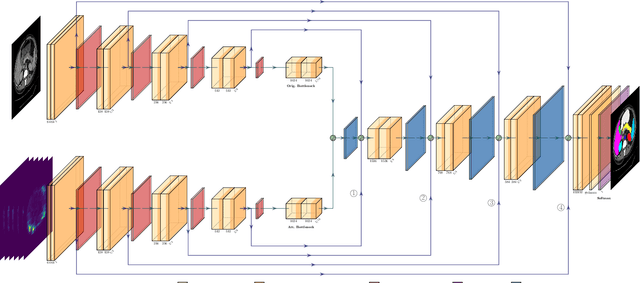
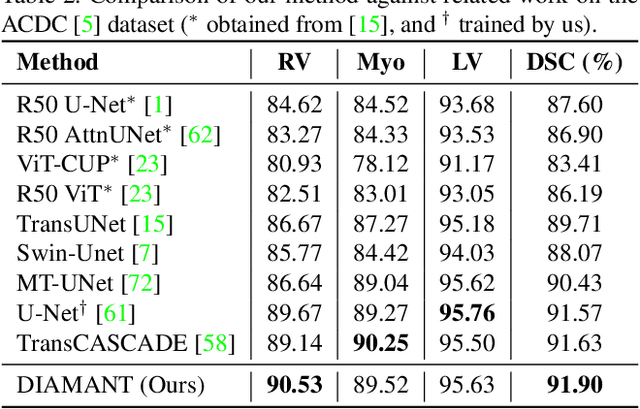
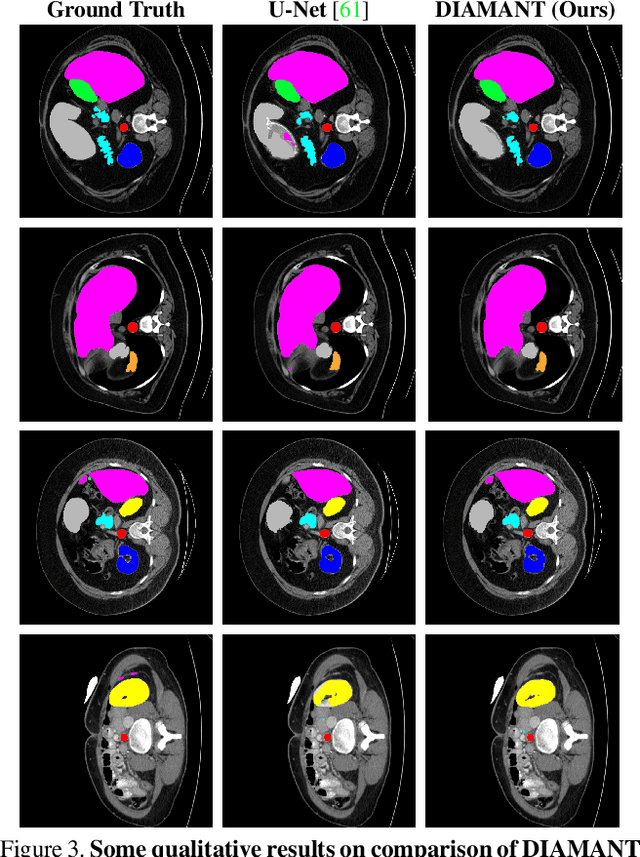
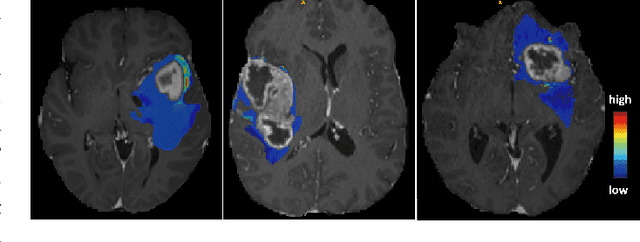
Although purely transformer-based architectures showed promising performance in many computer vision tasks, many hybrid models consisting of CNN and transformer blocks are introduced to fit more specialized tasks. Nevertheless, despite the performance gain of both pure and hybrid transformer-based architectures compared to CNNs in medical imaging segmentation, their high training cost and complexity make it challenging to use them in real scenarios. In this work, we propose simple architectures based on purely convolutional layers, and show that by just taking advantage of the attention map visualizations obtained from a self-supervised pretrained vision transformer network (e.g., DINO) one can outperform complex transformer-based networks with much less computation costs. The proposed architecture is composed of two encoder branches with the original image as input in one branch and the attention map visualizations of the same image from multiple self-attention heads from a pre-trained DINO model (as multiple channels) in the other branch. The results of our experiments on two publicly available medical imaging datasets show that the proposed pipeline outperforms U-Net and the state-of-the-art medical image segmentation models.
Open-Source Skull Reconstruction with MONAI
Nov 25, 2022Jianning Li, André Ferreira, Behrus Puladi, Victor Alves, Michael Kamp, Moon-Sung Kim, Felix Nensa, Jens Kleesiek, Seyed-Ahmad Ahmadi, Jan Egger
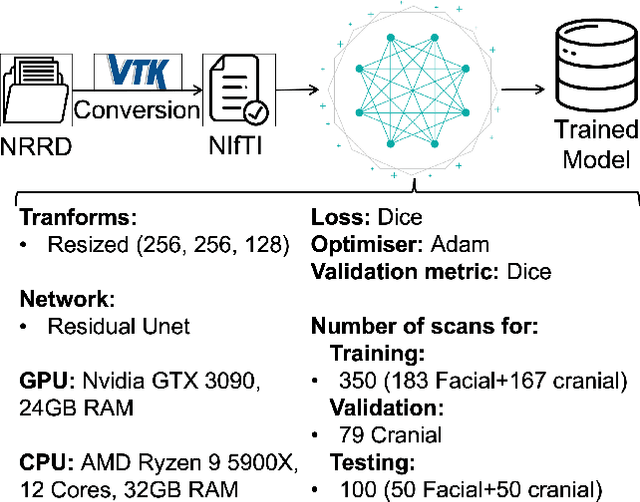
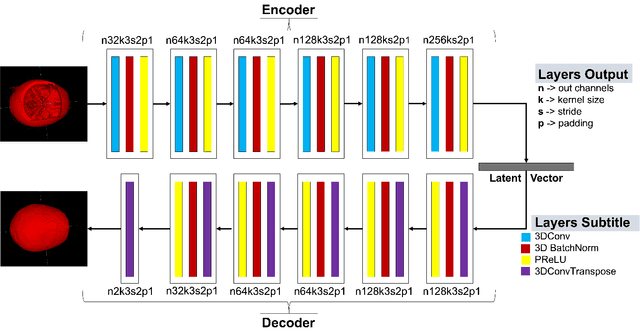
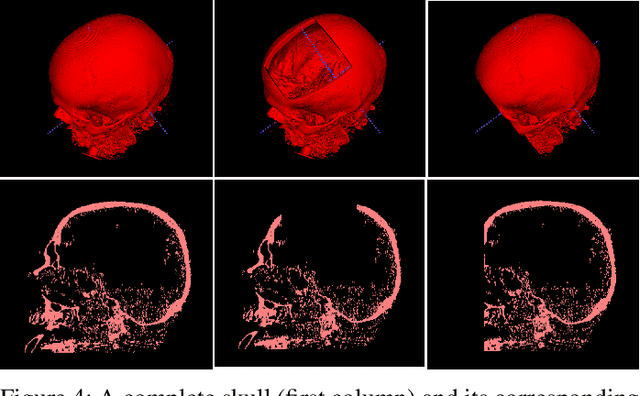
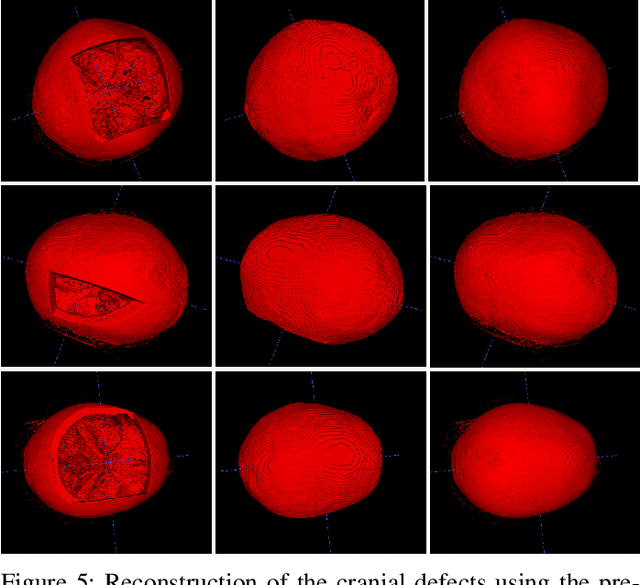
We present a deep learning-based approach for skull reconstruction for MONAI, which has been pre-trained on the MUG500+ skull dataset. The implementation follows the MONAI contribution guidelines, hence, it can be easily tried out and used, and extended by MONAI users. The primary goal of this paper lies in the investigation of open-sourcing codes and pre-trained deep learning models under the MONAI framework. Nowadays, open-sourcing software, especially (pre-trained) deep learning models, has become increasingly important. Over the years, medical image analysis experienced a tremendous transformation. Over a decade ago, algorithms had to be implemented and optimized with low-level programming languages, like C or C++, to run in a reasonable time on a desktop PC, which was not as powerful as today's computers. Nowadays, users have high-level scripting languages like Python, and frameworks like PyTorch and TensorFlow, along with a sea of public code repositories at hand. As a result, implementations that had thousands of lines of C or C++ code in the past, can now be scripted with a few lines and in addition executed in a fraction of the time. To put this even on a higher level, the Medical Open Network for Artificial Intelligence (MONAI) framework tailors medical imaging research to an even more convenient process, which can boost and push the whole field. The MONAI framework is a freely available, community-supported, open-source and PyTorch-based framework, that also enables to provide research contributions with pre-trained models to others. Codes and pre-trained weights for skull reconstruction are publicly available at: https://github.com/Project-MONAI/research-contributions/tree/master/SkullRec
Training β-VAE by Aggregating a Learned Gaussian Posterior with a Decoupled Decoder
Sep 29, 2022Jianning Li, Jana Fragemann, Seyed-Ahmad Ahmadi, Jens Kleesiek, Jan Egger
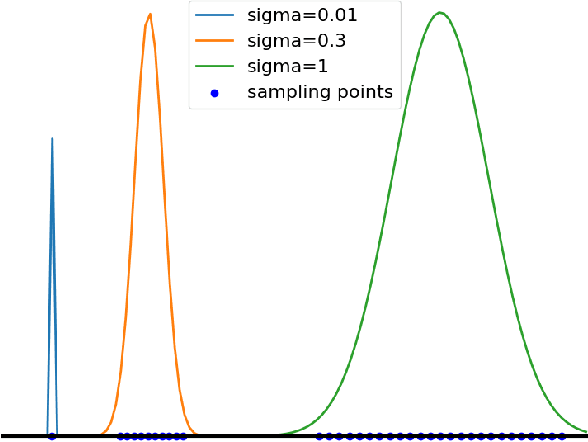
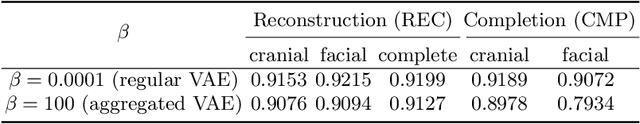
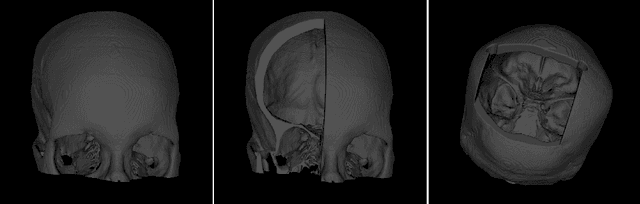
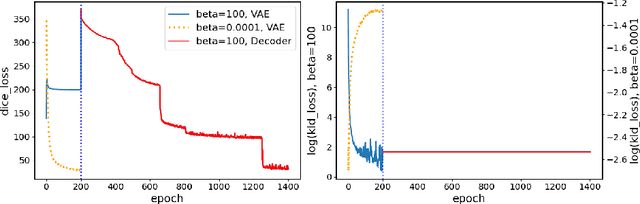
The reconstruction loss and the Kullback-Leibler divergence (KLD) loss in a variational autoencoder (VAE) often play antagonistic roles, and tuning the weight of the KLD loss in $\beta$-VAE to achieve a balance between the two losses is a tricky and dataset-specific task. As a result, current practices in VAE training often result in a trade-off between the reconstruction fidelity and the continuity$/$disentanglement of the latent space, if the weight $\beta$ is not carefully tuned. In this paper, we present intuitions and a careful analysis of the antagonistic mechanism of the two losses, and propose, based on the insights, a simple yet effective two-stage method for training a VAE. Specifically, the method aggregates a learned Gaussian posterior $z \sim q_{\theta} (z|x)$ with a decoder decoupled from the KLD loss, which is trained to learn a new conditional distribution $p_{\phi} (x|z)$ of the input data $x$. Experimentally, we show that the aggregated VAE maximally satisfies the Gaussian assumption about the latent space, while still achieves a reconstruction error comparable to when the latent space is only loosely regularized by $\mathcal{N}(\mathbf{0},I)$. The proposed approach does not require hyperparameter (i.e., the KLD weight $\beta$) tuning given a specific dataset as required in common VAE training practices. We evaluate the method using a medical dataset intended for 3D skull reconstruction and shape completion, and the results indicate promising generative capabilities of the VAE trained using the proposed method. Besides, through guided manipulation of the latent variables, we establish a connection between existing autoencoder (AE)-based approaches and generative approaches, such as VAE, for the shape completion problem. Codes and pre-trained weights are available at https://github.com/Jianningli/skullVAE
Graph-in-Graph (GiG): Learning interpretable latent graphs in non-Euclidean domain for biological and healthcare applications
Apr 01, 2022Kamilia Mullakaeva, Luca Cosmo, Anees Kazi, Seyed-Ahmad Ahmadi, Nassir Navab, Michael M. Bronstein
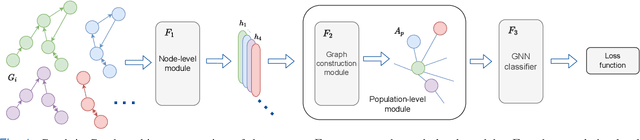
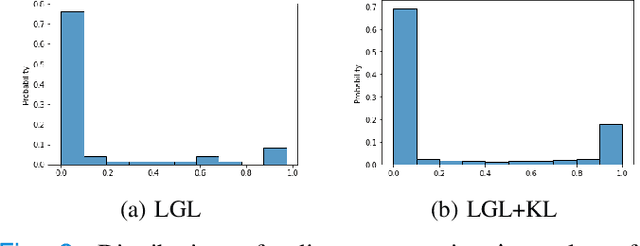
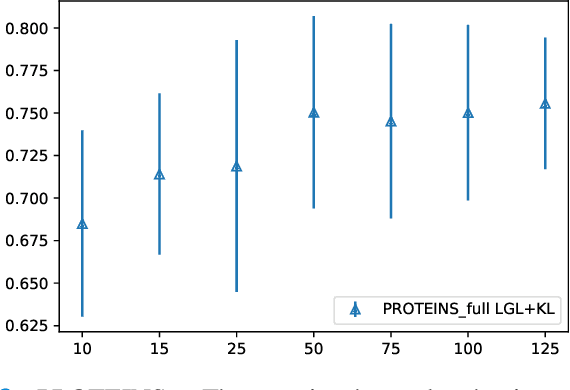
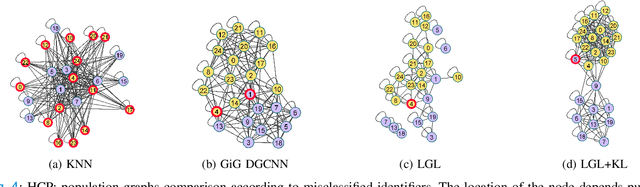
Graphs are a powerful tool for representing and analyzing unstructured, non-Euclidean data ubiquitous in the healthcare domain. Two prominent examples are molecule property prediction and brain connectome analysis. Importantly, recent works have shown that considering relationships between input data samples have a positive regularizing effect for the downstream task in healthcare applications. These relationships are naturally modeled by a (possibly unknown) graph structure between input samples. In this work, we propose Graph-in-Graph (GiG), a neural network architecture for protein classification and brain imaging applications that exploits the graph representation of the input data samples and their latent relation. We assume an initially unknown latent-graph structure between graph-valued input data and propose to learn end-to-end a parametric model for message passing within and across input graph samples, along with the latent structure connecting the input graphs. Further, we introduce a degree distribution loss that helps regularize the predicted latent relationships structure. This regularization can significantly improve the downstream task. Moreover, the obtained latent graph can represent patient population models or networks of molecule clusters, providing a level of interpretability and knowledge discovery in the input domain of particular value in healthcare.
Simultaneous imputation and disease classification in incomplete medical datasets using Multigraph Geometric Matrix Completion (MGMC)
May 14, 2020Gerome Vivar, Anees Kazi, Hendrik Burwinkel, Andreas Zwergal, Nassir Navab, Seyed-Ahmad Ahmadi
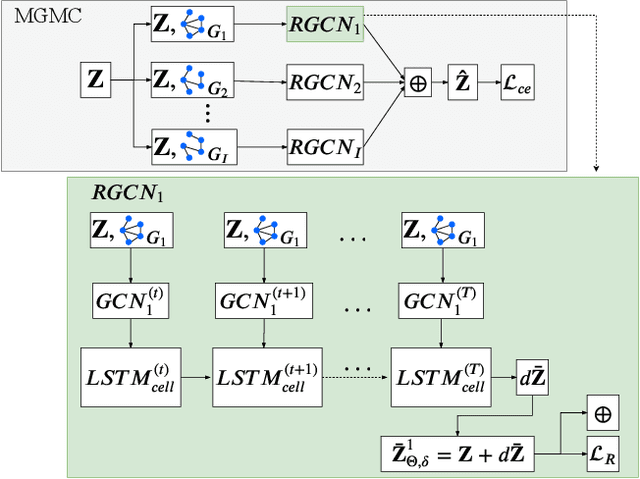
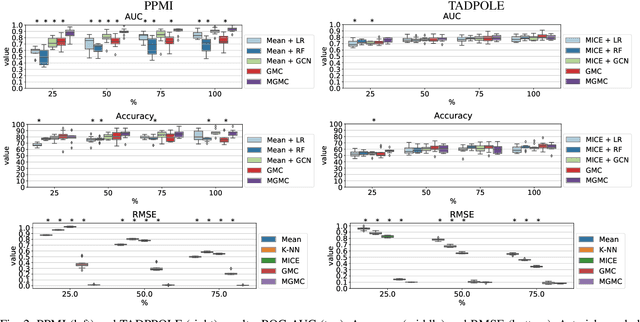
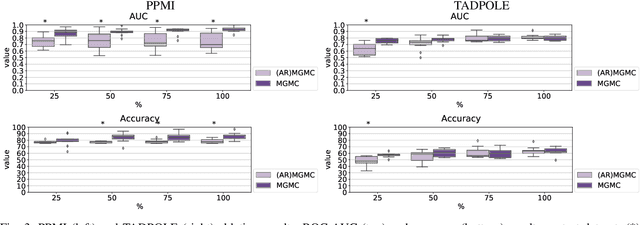
Large-scale population-based studies in medicine are a key resource towards better diagnosis, monitoring, and treatment of diseases. They also serve as enablers of clinical decision support systems, in particular Computer Aided Diagnosis (CADx) using machine learning (ML). Numerous ML approaches for CADx have been proposed in literature. However, these approaches assume full data availability, which is not always feasible in clinical data. To account for missing data, incomplete data samples are either removed or imputed, which could lead to data bias and may negatively affect classification performance. As a solution, we propose an end-to-end learning of imputation and disease prediction of incomplete medical datasets via Multigraph Geometric Matrix Completion (MGMC). MGMC uses multiple recurrent graph convolutional networks, where each graph represents an independent population model based on a key clinical meta-feature like age, sex, or cognitive function. Graph signal aggregation from local patient neighborhoods, combined with multigraph signal fusion via self-attention, has a regularizing effect on both matrix reconstruction and classification performance. Our proposed approach is able to impute class relevant features as well as perform accurate classification on two publicly available medical datasets. We empirically show the superiority of our proposed approach in terms of classification and imputation performance when compared with state-of-the-art approaches. MGMC enables disease prediction in multimodal and incomplete medical datasets. These findings could serve as baseline for future CADx approaches which utilize incomplete datasets.
Domain-specific loss design for unsupervised physical training: A new approach to modeling medical ML solutions
May 09, 2020Hendrik Burwinkel, Holger Matz, Stefan Saur, Christoph Hauger, Ayse Mine Evren, Nino Hirnschall, Oliver Findl, Nassir Navab, Seyed-Ahmad Ahmadi
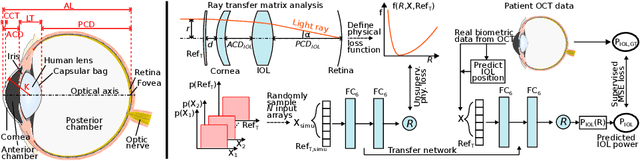
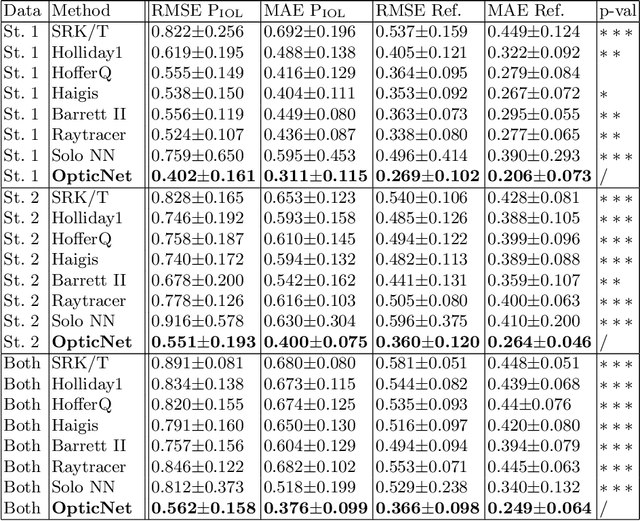
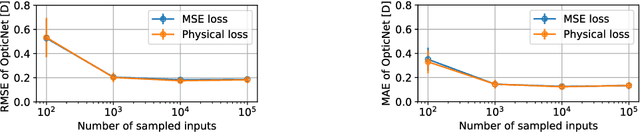
Today, cataract surgery is the most frequently performed ophthalmic surgery in the world. The cataract, a developing opacity of the human eye lens, constitutes the world's most frequent cause for blindness. During surgery, the lens is removed and replaced by an artificial intraocular lens (IOL). To prevent patients from needing strong visual aids after surgery, a precise prediction of the optical properties of the inserted IOL is crucial. There has been lots of activity towards developing methods to predict these properties from biometric eye data obtained by OCT devices, recently also by employing machine learning. They consider either only biometric data or physical models, but rarely both, and often neglect the IOL geometry. In this work, we propose OpticNet, a novel optical refraction network, loss function, and training scheme which is unsupervised, domain-specific, and physically motivated. We derive a precise light propagation eye model using single-ray raytracing and formulate a differentiable loss function that back-propagates physical gradients into the network. Further, we propose a new transfer learning procedure, which allows unsupervised training on the physical model and fine-tuning of the network on a cohort of real IOL patient cases. We show that our network is not only superior to systems trained with standard procedures but also that our method outperforms the current state of the art in IOL calculation when compared on two biometric data sets.
Decision Support for Intoxication Prediction Using Graph Convolutional Networks
May 02, 2020Hendrik Burwinkel, Matthias Keicher, David Bani-Harouni, Tobias Zellner, Florian Eyer, Nassir Navab, Seyed-Ahmad Ahmadi
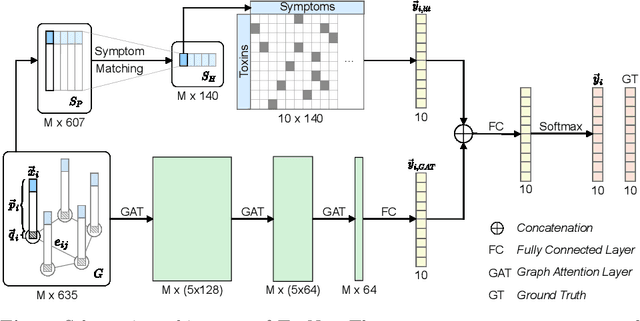
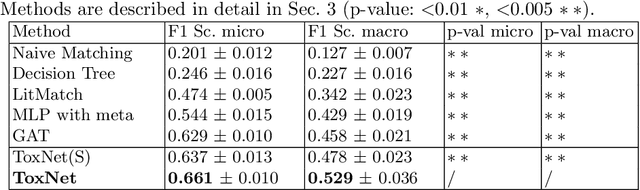
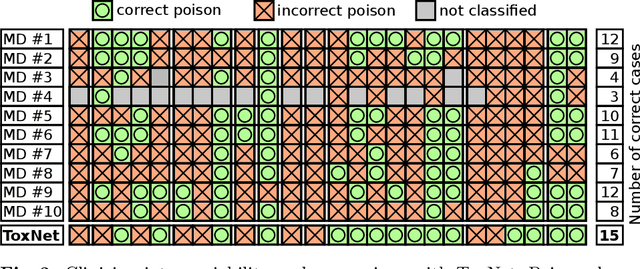
Every day, poison control centers (PCC) are called for immediate classification and treatment recommendations if an acute intoxication is suspected. Due to the time-sensitive nature of these cases, doctors are required to propose a correct diagnosis and intervention within a minimal time frame. Usually the toxin is known and recommendations can be made accordingly. However, in challenging cases only symptoms are mentioned and doctors have to rely on their clinical experience. Medical experts and our analyses of a regional dataset of intoxication records provide evidence that this is challenging, since occurring symptoms may not always match the textbook description due to regional distinctions, inter-rater variance, and institutional workflow. Computer-aided diagnosis (CADx) can provide decision support, but approaches so far do not consider additional information of the reported cases like age or gender, despite their potential value towards a correct diagnosis. In this work, we propose a new machine learning based CADx method which fuses symptoms and meta information of the patients using graph convolutional networks. We further propose a novel symptom matching method that allows the effective incorporation of prior knowledge into the learning process and evidently stabilizes the poison prediction. We validate our method against 10 medical doctors with different experience diagnosing intoxication cases for 10 different toxins from the PCC in Munich and show our method's superiority in performance for poison prediction.
Peri-Diagnostic Decision Support Through Cost-Efficient Feature Acquisition at Test-Time
Mar 31, 2020Gerome Vivar, Kamilia Mullakaeva, Andreas Zwergal, Nassir Navab, Seyed-Ahmad Ahmadi
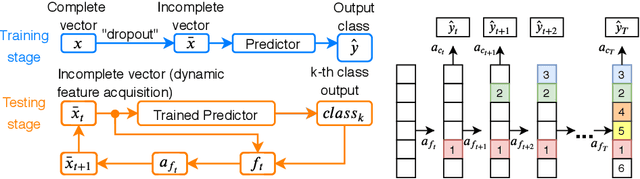
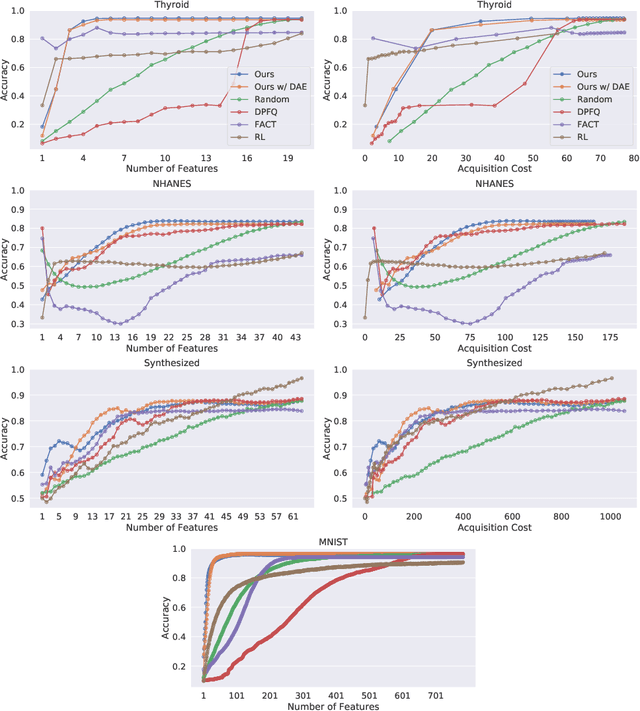
Computer-aided diagnosis (CADx) algorithms in medicine provide patient-specific decision support for physicians. These algorithms are usually applied after full acquisition of high-dimensional multimodal examination data, and often assume feature-completeness. This, however, is rarely the case due to examination costs, invasiveness, or a lack of indication. A sub-problem in CADx, which to our knowledge has received very little attention among the CADx community so far, is to guide the physician during the entire peri-diagnostic workflow, including the acquisition stage. We model the following question, asked from a physician's perspective: ''Given the evidence collected so far, which examination should I perform next, in order to achieve the most accurate and efficient diagnostic prediction?''. In this work, we propose a novel approach which is enticingly simple: use dropout at the input layer, and integrated gradients of the trained network at test-time to attribute feature importance dynamically. We validate and explain the effectiveness of our proposed approach using two public medical and two synthetic datasets. Results show that our proposed approach is more cost- and feature-efficient than prior approaches and achieves a higher overall accuracy. This directly translates to less unnecessary examinations for patients, and a quicker, less costly and more accurate decision support for the physician.
Latent Patient Network Learning for Automatic Diagnosis
Mar 27, 2020Luca Cosmo, Anees Kazi, Seyed-Ahmad Ahmadi, Nassir Navab, Michael Bronstein
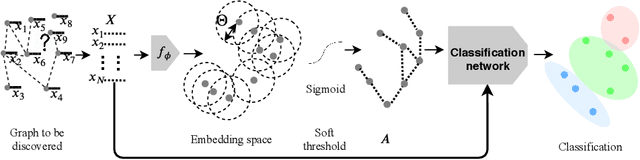
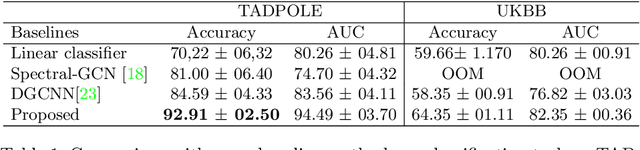
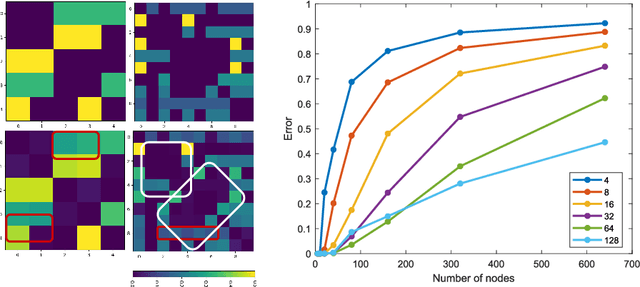
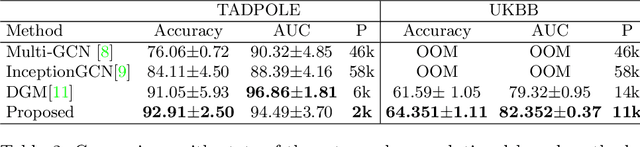
Recently, Graph Convolutional Networks (GCNs) has proven to be a powerful machine learning tool for Computer Aided Diagnosis (CADx) and disease prediction. A key component in these models is to build a population graph, where the graph adjacency matrix represents pair-wise patient similarities. Until now, the similarity metrics have been defined manually, usually based on meta-features like demographics or clinical scores. The definition of the metric, however, needs careful tuning, as GCNs are very sensitive to the graph structure. In this paper, we demonstrate for the first time in the CADx domain that it is possible to learn a single, optimal graph towards the GCN's downstream task of disease classification. To this end, we propose a novel, end-to-end trainable graph learning architecture for dynamic and localized graph pruning. Unlike commonly employed spectral GCN approaches, our GCN is spatial and inductive, and can thus infer previously unseen patients as well. We demonstrate significant classification improvements with our learned graph on two CADx problems in medicine. We further explain and visualize this result using an artificial dataset, underlining the importance of graph learning for more accurate and robust inference with GCNs in medical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge