Jan Egger
Enhancing Privacy: The Utility of Stand-Alone Synthetic CT and MRI for Tumor and Bone Segmentation
Jun 13, 2025Abstract:AI requires extensive datasets, while medical data is subject to high data protection. Anonymization is essential, but poses a challenge for some regions, such as the head, as identifying structures overlap with regions of clinical interest. Synthetic data offers a potential solution, but studies often lack rigorous evaluation of realism and utility. Therefore, we investigate to what extent synthetic data can replace real data in segmentation tasks. We employed head and neck cancer CT scans and brain glioma MRI scans from two large datasets. Synthetic data were generated using generative adversarial networks and diffusion models. We evaluated the quality of the synthetic data using MAE, MS-SSIM, Radiomics and a Visual Turing Test (VTT) performed by 5 radiologists and their usefulness in segmentation tasks using DSC. Radiomics indicates high fidelity of synthetic MRIs, but fall short in producing highly realistic CT tissue, with correlation coefficient of 0.8784 and 0.5461 for MRI and CT tumors, respectively. DSC results indicate limited utility of synthetic data: tumor segmentation achieved DSC=0.064 on CT and 0.834 on MRI, while bone segmentation a mean DSC=0.841. Relation between DSC and correlation is observed, but is limited by the complexity of the task. VTT results show synthetic CTs' utility, but with limited educational applications. Synthetic data can be used independently for the segmentation task, although limited by the complexity of the structures to segment. Advancing generative models to better tolerate heterogeneous inputs and learn subtle details is essential for enhancing their realism and expanding their application potential.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Comparative Analysis of nnUNet and MedNeXt for Head and Neck Tumor Segmentation in MRI-guided Radiotherapy
Nov 22, 2024Abstract:Radiation therapy (RT) is essential in treating head and neck cancer (HNC), with magnetic resonance imaging(MRI)-guided RT offering superior soft tissue contrast and functional imaging. However, manual tumor segmentation is time-consuming and complex, and therfore remains a challenge. In this study, we present our solution as team TUMOR to the HNTS-MRG24 MICCAI Challenge which is focused on automated segmentation of primary gross tumor volumes (GTVp) and metastatic lymph node gross tumor volume (GTVn) in pre-RT and mid-RT MRI images. We utilized the HNTS-MRG2024 dataset, which consists of 150 MRI scans from patients diagnosed with HNC, including original and registered pre-RT and mid-RT T2-weighted images with corresponding segmentation masks for GTVp and GTVn. We employed two state-of-the-art models in deep learning, nnUNet and MedNeXt. For Task 1, we pretrained models on pre-RT registered and mid-RT images, followed by fine-tuning on original pre-RT images. For Task 2, we combined registered pre-RT images, registered pre-RT segmentation masks, and mid-RT data as a multi-channel input for training. Our solution for Task 1 achieved 1st place in the final test phase with an aggregated Dice Similarity Coefficient of 0.8254, and our solution for Task 2 ranked 8th with a score of 0.7005. The proposed solution is publicly available at Github Repository.
Brain Tumour Removing and Missing Modality Generation using 3D WDM
Nov 07, 2024



Abstract:This paper presents the second-placed solution for task 8 and the participation solution for task 7 of BraTS 2024. The adoption of automated brain analysis algorithms to support clinical practice is increasing. However, many of these algorithms struggle with the presence of brain lesions or the absence of certain MRI modalities. The alterations in the brain's morphology leads to high variability and thus poor performance of predictive models that were trained only on healthy brains. The lack of information that is usually provided by some of the missing MRI modalities also reduces the reliability of the prediction models trained with all modalities. In order to improve the performance of these models, we propose the use of conditional 3D wavelet diffusion models. The wavelet transform enabled full-resolution image training and prediction on a GPU with 48 GB VRAM, without patching or downsampling, preserving all information for prediction. For the inpainting task of BraTS 2024, the use of a large and variable number of healthy masks and the stability and efficiency of the 3D wavelet diffusion model resulted in 0.007, 22.61 and 0.842 in the validation set and 0.07 , 22.8 and 0.91 in the testing set (MSE, PSNR and SSIM respectively). The code for these tasks is available at https://github.com/ShadowTwin41/BraTS_2023_2024_solutions.
Improved Multi-Task Brain Tumour Segmentation with Synthetic Data Augmentation
Nov 07, 2024



Abstract:This paper presents the winning solution of task 1 and the third-placed solution of task 3 of the BraTS challenge. The use of automated tools in clinical practice has increased due to the development of more and more sophisticated and reliable algorithms. However, achieving clinical standards and developing tools for real-life scenarios is a major challenge. To this end, BraTS has organised tasks to find the most advanced solutions for specific purposes. In this paper, we propose the use of synthetic data to train state-of-the-art frameworks in order to improve the segmentation of adult gliomas in a post-treatment scenario, and the segmentation of meningioma for radiotherapy planning. Our results suggest that the use of synthetic data leads to more robust algorithms, although the synthetic data generation pipeline is not directly suited to the meningioma task. The code for these tasks is available at https://github.com/ShadowTwin41/BraTS_2023_2024_solutions.
Spacewalker: Traversing Representation Spaces for Fast Interactive Exploration and Annotation of Unstructured Data
Sep 25, 2024



Abstract:Unstructured data in industries such as healthcare, finance, and manufacturing presents significant challenges for efficient analysis and decision making. Detecting patterns within this data and understanding their impact is critical but complex without the right tools. Traditionally, these tasks relied on the expertise of data analysts or labor-intensive manual reviews. In response, we introduce Spacewalker, an interactive tool designed to explore and annotate data across multiple modalities. Spacewalker allows users to extract data representations and visualize them in low-dimensional spaces, enabling the detection of semantic similarities. Through extensive user studies, we assess Spacewalker's effectiveness in data annotation and integrity verification. Results show that the tool's ability to traverse latent spaces and perform multi-modal queries significantly enhances the user's capacity to quickly identify relevant data. Moreover, Spacewalker allows for annotation speed-ups far superior to conventional methods, making it a promising tool for efficiently navigating unstructured data and improving decision making processes. The code of this work is open-source and can be found at: https://github.com/code-lukas/Spacewalker
CC-DCNet: Dynamic Convolutional Neural Network with Contrastive Constraints for Identifying Lung Cancer Subtypes on Multi-modality Images
Jul 18, 2024



Abstract:The accurate diagnosis of pathological subtypes of lung cancer is of paramount importance for follow-up treatments and prognosis managements. Assessment methods utilizing deep learning technologies have introduced novel approaches for clinical diagnosis. However, the majority of existing models rely solely on single-modality image input, leading to limited diagnostic accuracy. To this end, we propose a novel deep learning network designed to accurately classify lung cancer subtype with multi-dimensional and multi-modality images, i.e., CT and pathological images. The strength of the proposed model lies in its ability to dynamically process both paired CT-pathological image sets as well as independent CT image sets, and consequently optimize the pathology-related feature extractions from CT images. This adaptive learning approach enhances the flexibility in processing multi-dimensional and multi-modality datasets and results in performance elevating in the model testing phase. We also develop a contrastive constraint module, which quantitatively maps the cross-modality associations through network training, and thereby helps to explore the "gold standard" pathological information from the corresponding CT scans. To evaluate the effectiveness, adaptability, and generalization ability of our model, we conducted extensive experiments on a large-scale multi-center dataset and compared our model with a series of state-of-the-art classification models. The experimental results demonstrated the superiority of our model for lung cancer subtype classification, showcasing significant improvements in accuracy metrics such as ACC, AUC, and F1-score.
Deep Dive into MRI: Exploring Deep Learning Applications in 0.55T and 7T MRI
Jul 01, 2024


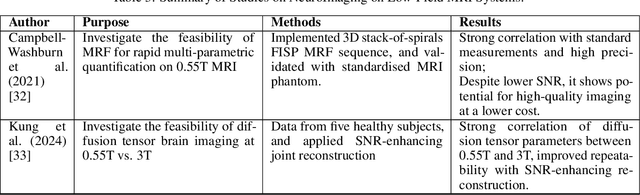
Abstract:The development of magnetic resonance imaging (MRI) for medical imaging has provided a leap forward in diagnosis, providing a safe, non-invasive alternative to techniques involving ionising radiation exposure for diagnostic purposes. It was described by Block and Purcel in 1946, and it was not until 1980 that the first clinical application of MRI became available. Since that time the MRI has gone through many advances and has altered the way diagnosing procedures are performed. Due to its ability to improve constantly, MRI has become a commonly used practice among several specialisations in medicine. Particularly starting 0.55T and 7T MRI technologies have pointed out enhanced preservation of image detail and advanced tissue characterisation. This review examines the integration of deep learning (DL) techniques into these MRI modalities, disseminating and exploring the study applications. It highlights how DL contributes to 0.55T and 7T MRI data, showcasing the potential of DL in improving and refining these technologies. The review ends with a brief overview of how MRI technology will evolve in the coming years.
Deep Learning-based Point Cloud Registration for Augmented Reality-guided Surgery
May 06, 2024Abstract:Point cloud registration aligns 3D point clouds using spatial transformations. It is an important task in computer vision, with applications in areas such as augmented reality (AR) and medical imaging. This work explores the intersection of two research trends: the integration of AR into image-guided surgery and the use of deep learning for point cloud registration. The main objective is to evaluate the feasibility of applying deep learning-based point cloud registration methods for image-to-patient registration in augmented reality-guided surgery. We created a dataset of point clouds from medical imaging and corresponding point clouds captured with a popular AR device, the HoloLens 2. We evaluate three well-established deep learning models in registering these data pairs. While we find that some deep learning methods show promise, we show that a conventional registration pipeline still outperforms them on our challenging dataset.
Deep Medial Voxels: Learned Medial Axis Approximations for Anatomical Shape Modeling
Mar 18, 2024Abstract:Shape reconstruction from imaging volumes is a recurring need in medical image analysis. Common workflows start with a segmentation step, followed by careful post-processing and,finally, ad hoc meshing algorithms. As this sequence can be timeconsuming, neural networks are trained to reconstruct shapes through template deformation. These networks deliver state-ofthe-art results without manual intervention, but, so far, they have primarily been evaluated on anatomical shapes with little topological variety between individuals. In contrast, other works favor learning implicit shape models, which have multiple benefits for meshing and visualization. Our work follows this direction by introducing deep medial voxels, a semi-implicit representation that faithfully approximates the topological skeleton from imaging volumes and eventually leads to shape reconstruction via convolution surfaces. Our reconstruction technique shows potential for both visualization and computer simulations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge