Rongkun Zhu
From Orthomosaics to Raw UAV Imagery: Enhancing Palm Detection and Crown-Center Localization
Sep 15, 2025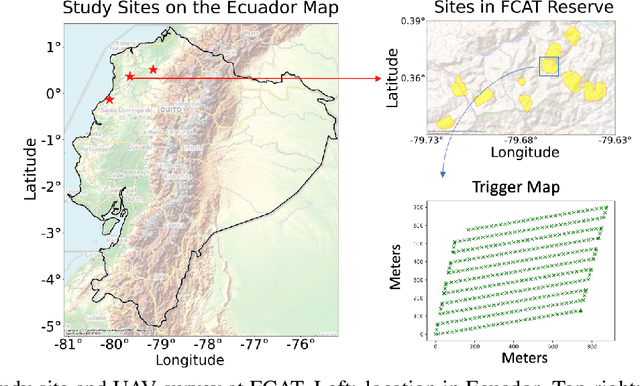
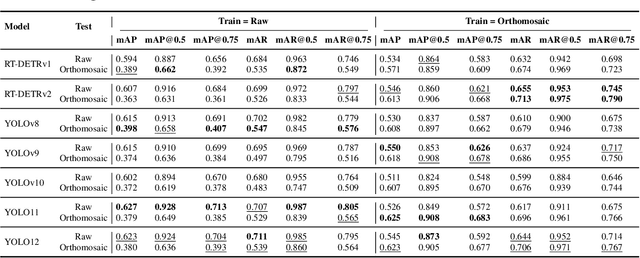
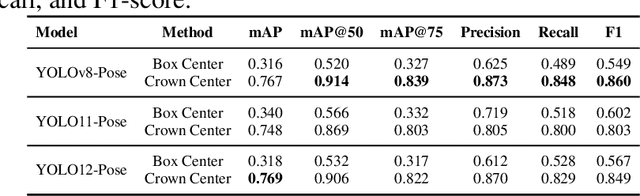

Abstract:Accurate mapping of individual trees is essential for ecological monitoring and forest management. Orthomosaic imagery from unmanned aerial vehicles (UAVs) is widely used, but stitching artifacts and heavy preprocessing limit its suitability for field deployment. This study explores the use of raw UAV imagery for palm detection and crown-center localization in tropical forests. Two research questions are addressed: (1) how detection performance varies across orthomosaic and raw imagery, including within-domain and cross-domain transfer, and (2) to what extent crown-center annotations improve localization accuracy beyond bounding-box centroids. Using state-of-the-art detectors and keypoint models, we show that raw imagery yields superior performance in deployment-relevant scenarios, while orthomosaics retain value for robust cross-domain generalization. Incorporating crown-center annotations in training further improves localization and provides precise tree positions for downstream ecological analyses. These findings offer practical guidance for UAV-based biodiversity and conservation monitoring.
U2-BENCH: Benchmarking Large Vision-Language Models on Ultrasound Understanding
May 23, 2025Abstract:Ultrasound is a widely-used imaging modality critical to global healthcare, yet its interpretation remains challenging due to its varying image quality on operators, noises, and anatomical structures. Although large vision-language models (LVLMs) have demonstrated impressive multimodal capabilities across natural and medical domains, their performance on ultrasound remains largely unexplored. We introduce U2-BENCH, the first comprehensive benchmark to evaluate LVLMs on ultrasound understanding across classification, detection, regression, and text generation tasks. U2-BENCH aggregates 7,241 cases spanning 15 anatomical regions and defines 8 clinically inspired tasks, such as diagnosis, view recognition, lesion localization, clinical value estimation, and report generation, across 50 ultrasound application scenarios. We evaluate 20 state-of-the-art LVLMs, both open- and closed-source, general-purpose and medical-specific. Our results reveal strong performance on image-level classification, but persistent challenges in spatial reasoning and clinical language generation. U2-BENCH establishes a rigorous and unified testbed to assess and accelerate LVLM research in the uniquely multimodal domain of medical ultrasound imaging.
Detection and Geographic Localization of Natural Objects in the Wild: A Case Study on Palms
Feb 18, 2025



Abstract:Palms are ecologically and economically indicators of tropical forest health, biodiversity, and human impact that support local economies and global forest product supply chains. While palm detection in plantations is well-studied, efforts to map naturally occurring palms in dense forests remain limited by overlapping crowns, uneven shading, and heterogeneous landscapes. We develop PRISM (Processing, Inference, Segmentation, and Mapping), a flexible pipeline for detecting and localizing palms in dense tropical forests using large orthomosaic images. Orthomosaics are created from thousands of aerial images and spanning several to hundreds of gigabytes. Our contributions are threefold. First, we construct a large UAV-derived orthomosaic dataset collected across 21 ecologically diverse sites in western Ecuador, annotated with 8,830 bounding boxes and 5,026 palm center points. Second, we evaluate multiple state-of-the-art object detectors based on efficiency and performance, integrating zero-shot SAM 2 as the segmentation backbone, and refining the results for precise geographic mapping. Third, we apply calibration methods to align confidence scores with IoU and explore saliency maps for feature explainability. Though optimized for palms, PRISM is adaptable for identifying other natural objects, such as eastern white pines. Future work will explore transfer learning for lower-resolution datasets (0.5 to 1m).
Real-Time Localization and Bimodal Point Pattern Analysis of Palms Using UAV Imagery
Oct 14, 2024



Abstract:Understanding the spatial distribution of palms within tropical forests is essential for effective ecological monitoring, conservation strategies, and the sustainable integration of natural forest products into local and global supply chains. However, the analysis of remotely sensed data in these environments faces significant challenges, such as overlapping palm and tree crowns, uneven shading across the canopy surface, and the heterogeneous nature of the forest landscapes, which often affect the performance of palm detection and segmentation algorithms. To overcome these issues, we introduce PalmDSNet, a deep learning framework for real-time detection, segmentation, and counting of canopy palms. Additionally, we employ a bimodal reproduction algorithm that simulates palm spatial propagation to further enhance the understanding of these point patterns using PalmDSNet's results. We used UAV-captured imagery to create orthomosaics from 21 sites across western Ecuadorian tropical forests, covering a gradient from the everwet Choc\'o forests near Colombia to the drier forests of southwestern Ecuador. Expert annotations were used to create a comprehensive dataset, including 7,356 bounding boxes on image patches and 7,603 palm centers across five orthomosaics, encompassing a total area of 449 hectares. By combining PalmDSNet with the bimodal reproduction algorithm, which optimizes parameters for both local and global spatial variability, we effectively simulate the spatial distribution of palms in diverse and dense tropical environments, validating its utility for advanced applications in tropical forest monitoring and remote sensing analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge