Paul Klein
EchoApex: A General-Purpose Vision Foundation Model for Echocardiography
Oct 14, 2024



Abstract:Quantitative evaluation of echocardiography is essential for precise assessment of cardiac condition, monitoring disease progression, and guiding treatment decisions. The diverse nature of echo images, including variations in probe types, manufacturers, and pathologies, poses challenges for developing artificial intelligent models that can generalize across different clinical practice. We introduce EchoApex, the first general-purpose vision foundation model echocardiography with applications on a variety of clinical practice. Leveraging self-supervised learning, EchoApex is pretrained on over 20 million echo images from 11 clinical centres. By incorporating task-specific decoders and adapter modules, we demonstrate the effectiveness of EchoApex on 4 different kind of clinical applications with 28 sub-tasks, including view classification, interactive structure segmentation, left ventricle hypertrophy detection and automated ejection fraction estimation from view sequences. Compared to state-of-the-art task-specific models, EchoApex attains improved performance with a unified image encoding architecture, demonstrating the benefits of model pretraining at scale with in-domain data. Furthermore, EchoApex illustrates the potential for developing a general-purpose vision foundation model tailored specifically for echocardiography, capable of addressing a diverse range of clinical applications with high efficiency and efficacy.
AI-driven View Guidance System in Intra-cardiac Echocardiography Imaging
Sep 26, 2024



Abstract:Intra-cardiac Echocardiography (ICE) is a crucial imaging modality used in electrophysiology (EP) and structural heart disease (SHD) interventions, providing real-time, high-resolution views from within the heart. Despite its advantages, effective manipulation of the ICE catheter requires significant expertise, which can lead to inconsistent outcomes, particularly among less experienced operators. To address this challenge, we propose an AI-driven closed-loop view guidance system with human-in-the-loop feedback, designed to assist users in navigating ICE imaging without requiring specialized knowledge. Our method models the relative position and orientation vectors between arbitrary views and clinically defined ICE views in a spatial coordinate system, guiding users on how to manipulate the ICE catheter to transition from the current view to the desired view over time. Operating in a closed-loop configuration, the system continuously predicts and updates the necessary catheter manipulations, ensuring seamless integration into existing clinical workflows. The effectiveness of the proposed system is demonstrated through a simulation-based evaluation, achieving an 89% success rate with the 6532 test dataset, highlighting its potential to improve the accuracy and efficiency of ICE imaging procedures.
$Γ$-VAE: Curvature regularized variational autoencoders for uncovering emergent low dimensional geometric structure in high dimensional data
Mar 02, 2024



Abstract:Natural systems with emergent behaviors often organize along low-dimensional subsets of high-dimensional spaces. For example, despite the tens of thousands of genes in the human genome, the principled study of genomics is fruitful because biological processes rely on coordinated organization that results in lower dimensional phenotypes. To uncover this organization, many nonlinear dimensionality reduction techniques have successfully embedded high-dimensional data into low-dimensional spaces by preserving local similarities between data points. However, the nonlinearities in these methods allow for too much curvature to preserve general trends across multiple non-neighboring data clusters, thereby limiting their interpretability and generalizability to out-of-distribution data. Here, we address both of these limitations by regularizing the curvature of manifolds generated by variational autoencoders, a process we coin ``$\Gamma$-VAE''. We demonstrate its utility using two example data sets: bulk RNA-seq from the The Cancer Genome Atlas (TCGA) and the Genotype Tissue Expression (GTEx); and single cell RNA-seq from a lineage tracing experiment in hematopoietic stem cell differentiation. We find that the resulting regularized manifolds identify mesoscale structure associated with different cancer cell types, and accurately re-embed tissues from completely unseen, out-of distribution cancers as if they were originally trained on them. Finally, we show that preserving long-range relationships to differentiated cells separates undifferentiated cells -- which have not yet specialized -- according to their eventual fate. Broadly, we anticipate that regularizing the curvature of generative models will enable more consistent, predictive, and generalizable models in any high-dimensional system with emergent low-dimensional behavior.
Cardiac ultrasound simulation for autonomous ultrasound navigation
Feb 09, 2024Abstract:Ultrasound is well-established as an imaging modality for diagnostic and interventional purposes. However, the image quality varies with operator skills as acquiring and interpreting ultrasound images requires extensive training due to the imaging artefacts, the range of acquisition parameters and the variability of patient anatomies. Automating the image acquisition task could improve acquisition reproducibility and quality but training such an algorithm requires large amounts of navigation data, not saved in routine examinations. Thus, we propose a method to generate large amounts of ultrasound images from other modalities and from arbitrary positions, such that this pipeline can later be used by learning algorithms for navigation. We present a novel simulation pipeline which uses segmentations from other modalities, an optimized volumetric data representation and GPU-accelerated Monte Carlo path tracing to generate view-dependent and patient-specific ultrasound images. We extensively validate the correctness of our pipeline with a phantom experiment, where structures' sizes, contrast and speckle noise properties are assessed. Furthermore, we demonstrate its usability to train neural networks for navigation in an echocardiography view classification experiment by generating synthetic images from more than 1000 patients. Networks pre-trained with our simulations achieve significantly superior performance in settings where large real datasets are not available, especially for under-represented classes. The proposed approach allows for fast and accurate patient-specific ultrasound image generation, and its usability for training networks for navigation-related tasks is demonstrated.
Graph Cut Segmentation Methods Revisited with a Quantum Algorithm
Dec 07, 2018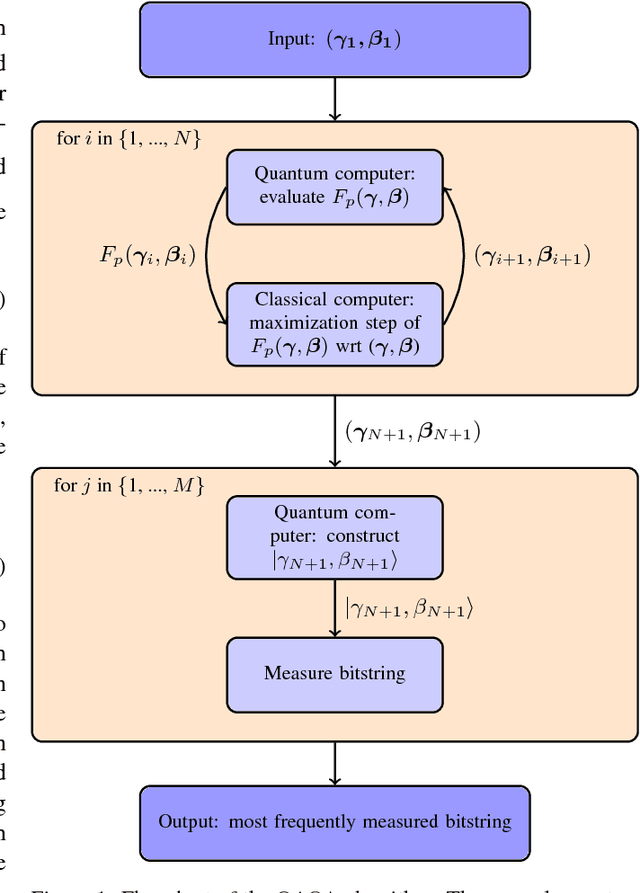
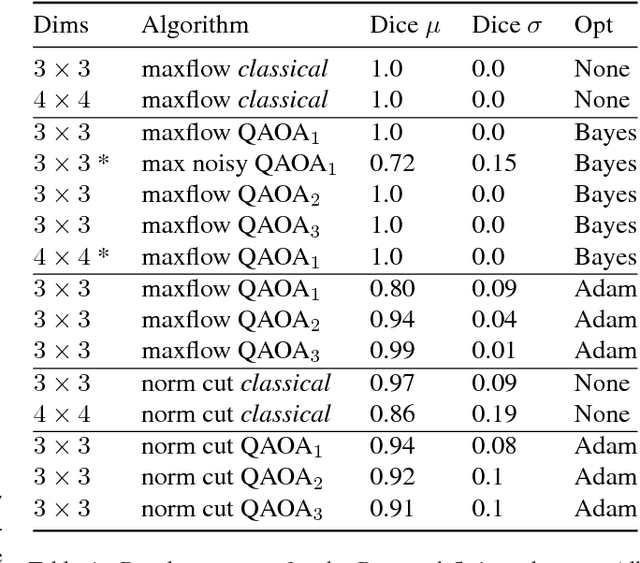
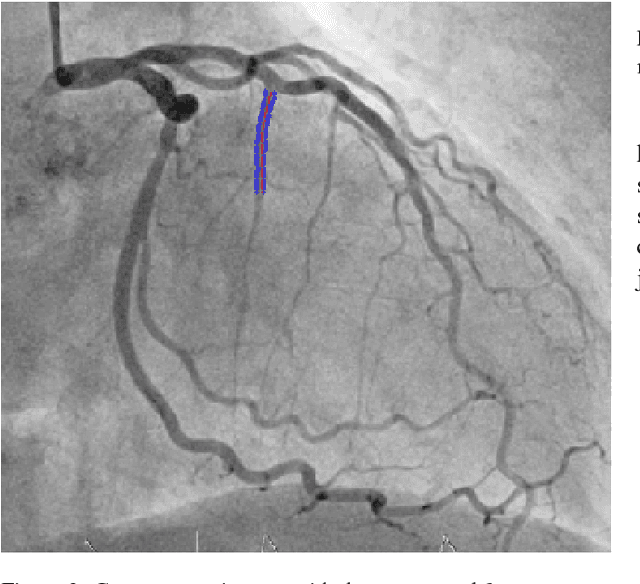
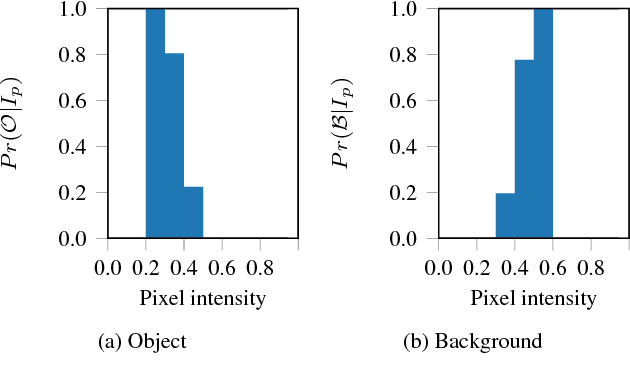
Abstract:The design and performance of computer vision algorithms are greatly influenced by the hardware on which they are implemented. CPUs, multi-core CPUs, FPGAs and GPUs have inspired new algorithms and enabled existing ideas to be realized. This is notably the case with GPUs, which has significantly changed the landscape of computer vision research through deep learning. As the end of Moores law approaches, researchers and hardware manufacturers are exploring alternative hardware computing paradigms. Quantum computers are a very promising alternative and offer polynomial or even exponential speed-ups over conventional computing for some problems. This paper presents a novel approach to image segmentation that uses new quantum computing hardware. Segmentation is formulated as a graph cut problem that can be mapped to the quantum approximation optimization algorithm (QAOA). This algorithm can be implemented on current and near-term quantum computers. Encouraging results are presented on artificial and medical imaging data. This represents an important, practical step towards leveraging quantum computers for computer vision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge