Parashkev Nachev
Bidirectional human-AI collaboration in brain tumour assessments improves both expert human and AI agent performance
Dec 13, 2025Abstract:The benefits of artificial intelligence (AI) human partnerships-evaluating how AI agents enhance expert human performance-are increasingly studied. Though rarely evaluated in healthcare, an inverse approach is possible: AI benefiting from the support of an expert human agent. Here, we investigate both human-AI clinical partnership paradigms in the magnetic resonance imaging-guided characterisation of patients with brain tumours. We reveal that human-AI partnerships improve accuracy and metacognitive ability not only for radiologists supported by AI, but also for AI agents supported by radiologists. Moreover, the greatest patient benefit was evident with an AI agent supported by a human one. Synergistic improvements in agent accuracy, metacognitive performance, and inter-rater agreement suggest that AI can create more capable, confident, and consistent clinical agents, whether human or model-based. Our work suggests that the maximal value of AI in healthcare could emerge not from replacing human intelligence, but from AI agents that routinely leverage and amplify it.
Generalizable automated ischaemic stroke lesion segmentation with vision transformers
Feb 10, 2025



Abstract:Ischaemic stroke, a leading cause of death and disability, critically relies on neuroimaging for characterising the anatomical pattern of injury. Diffusion-weighted imaging (DWI) provides the highest expressivity in ischemic stroke but poses substantial challenges for automated lesion segmentation: susceptibility artefacts, morphological heterogeneity, age-related comorbidities, time-dependent signal dynamics, instrumental variability, and limited labelled data. Current U-Net-based models therefore underperform, a problem accentuated by inadequate evaluation metrics that focus on mean performance, neglecting anatomical, subpopulation, and acquisition-dependent variability. Here, we present a high-performance DWI lesion segmentation tool addressing these challenges through optimized vision transformer-based architectures, integration of 3563 annotated lesions from multi-site data, and algorithmic enhancements, achieving state-of-the-art results. We further propose a novel evaluative framework assessing model fidelity, equity (across demographics and lesion subtypes), anatomical precision, and robustness to instrumental variability, promoting clinical and research utility. This work advances stroke imaging by reconciling model expressivity with domain-specific challenges and redefining performance benchmarks to prioritize equity and generalizability, critical for personalized medicine and mechanistic research.
Deep generative computed perfusion-deficit mapping of ischaemic stroke
Feb 03, 2025



Abstract:Focal deficits in ischaemic stroke result from impaired perfusion downstream of a critical vascular occlusion. While parenchymal lesions are traditionally used to predict clinical deficits, the underlying pattern of disrupted perfusion provides information upstream of the lesion, potentially yielding earlier predictive and localizing signals. Such perfusion maps can be derived from routine CT angiography (CTA) widely deployed in clinical practice. Analysing computed perfusion maps from 1,393 CTA-imaged-patients with acute ischaemic stroke, we use deep generative inference to localise neural substrates of NIHSS sub-scores. We show that our approach replicates known lesion-deficit relations without knowledge of the lesion itself and reveals novel neural dependents. The high achieved anatomical fidelity suggests acute CTA-derived computed perfusion maps may be of substantial clinical-and-scientific value in rich phenotyping of acute stroke. Using only hyperacute imaging, deep generative inference could power highly expressive models of functional anatomical relations in ischaemic stroke within the pre-interventional window.
UniBrain: A Unified Model for Cross-Subject Brain Decoding
Dec 27, 2024



Abstract:Brain decoding aims to reconstruct original stimuli from fMRI signals, providing insights into interpreting mental content. Current approaches rely heavily on subject-specific models due to the complex brain processing mechanisms and the variations in fMRI signals across individuals. Therefore, these methods greatly limit the generalization of models and fail to capture cross-subject commonalities. To address this, we present UniBrain, a unified brain decoding model that requires no subject-specific parameters. Our approach includes a group-based extractor to handle variable fMRI signal lengths, a mutual assistance embedder to capture cross-subject commonalities, and a bilevel feature alignment scheme for extracting subject-invariant features. We validate our UniBrain on the brain decoding benchmark, achieving comparable performance to current state-of-the-art subject-specific models with extremely fewer parameters. We also propose a generalization benchmark to encourage the community to emphasize cross-subject commonalities for more general brain decoding. Our code is available at https://github.com/xiaoyao3302/UniBrain.
Deep Generative Classification of Blood Cell Morphology
Aug 16, 2024



Abstract:Accurate classification of haematological cells is critical for diagnosing blood disorders, but presents significant challenges for machine automation owing to the complexity of cell morphology, heterogeneities of biological, pathological, and imaging characteristics, and the imbalance of cell type frequencies. We introduce CytoDiffusion, a diffusion-based classifier that effectively models blood cell morphology, combining accurate classification with robust anomaly detection, resistance to distributional shifts, interpretability, data efficiency, and superhuman uncertainty quantification. Our approach outperforms state-of-the-art discriminative models in anomaly detection (AUC 0.976 vs. 0.919), resistance to domain shifts (85.85% vs. 74.38% balanced accuracy), and performance in low-data regimes (95.88% vs. 94.95% balanced accuracy). Notably, our model generates synthetic blood cell images that are nearly indistinguishable from real images, as demonstrated by a Turing test in which expert haematologists achieved only 52.3% accuracy (95% CI: [50.5%, 54.2%]). Furthermore, we enhance model explainability through the generation of directly interpretable counterfactual heatmaps. Our comprehensive evaluation framework, encompassing these multiple performance dimensions, establishes a new benchmark for medical image analysis in haematology, ultimately enabling improved diagnostic accuracy in clinical settings. Our code is available at https://github.com/Deltadahl/CytoDiffusion.
Framework to generate perfusion map from CT and CTA images in patients with acute ischemic stroke: A longitudinal and cross-sectional study
Apr 05, 2024



Abstract:Stroke is a leading cause of disability and death. Effective treatment decisions require early and informative vascular imaging. 4D perfusion imaging is ideal but rarely available within the first hour after stroke, whereas plain CT and CTA usually are. Hence, we propose a framework to extract a predicted perfusion map (PPM) derived from CT and CTA images. In all eighteen patients, we found significantly high spatial similarity (with average Spearman's correlation = 0.7893) between our predicted perfusion map (PPM) and the T-max map derived from 4D-CTP. Voxelwise correlations between the PPM and National Institutes of Health Stroke Scale (NIHSS) subscores for L/R hand motor, gaze, and language on a large cohort of 2,110 subjects reliably mapped symptoms to expected infarct locations. Therefore our PPM could serve as an alternative for 4D perfusion imaging, if the latter is unavailable, to investigate blood perfusion in the first hours after hospital admission.
RAISE -- Radiology AI Safety, an End-to-end lifecycle approach
Nov 24, 2023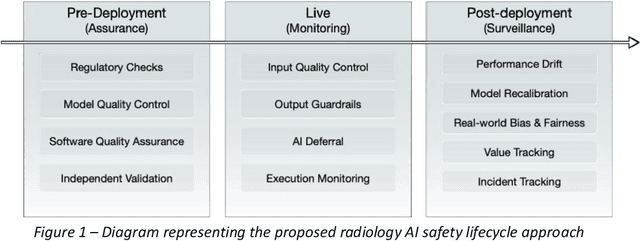
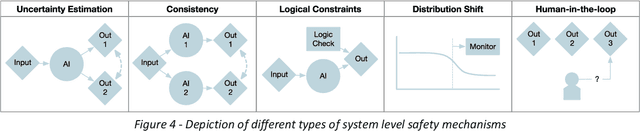
Abstract:The integration of AI into radiology introduces opportunities for improved clinical care provision and efficiency but it demands a meticulous approach to mitigate potential risks as with any other new technology. Beginning with rigorous pre-deployment evaluation and validation, the focus should be on ensuring models meet the highest standards of safety, effectiveness and efficacy for their intended applications. Input and output guardrails implemented during production usage act as an additional layer of protection, identifying and addressing individual failures as they occur. Continuous post-deployment monitoring allows for tracking population-level performance (data drift), fairness, and value delivery over time. Scheduling reviews of post-deployment model performance and educating radiologists about new algorithmic-driven findings is critical for AI to be effective in clinical practice. Recognizing that no single AI solution can provide absolute assurance even when limited to its intended use, the synergistic application of quality assurance at multiple levels - regulatory, clinical, technical, and ethical - is emphasized. Collaborative efforts between stakeholders spanning healthcare systems, industry, academia, and government are imperative to address the multifaceted challenges involved. Trust in AI is an earned privilege, contingent on a broad set of goals, among them transparently demonstrating that the AI adheres to the same rigorous safety, effectiveness and efficacy standards as other established medical technologies. By doing so, developers can instil confidence among providers and patients alike, enabling the responsible scaling of AI and the realization of its potential benefits. The roadmap presented herein aims to expedite the achievement of deployable, reliable, and safe AI in radiology.
Compressed representation of brain genetic transcription
Oct 24, 2023Abstract:The architecture of the brain is too complex to be intuitively surveyable without the use of compressed representations that project its variation into a compact, navigable space. The task is especially challenging with high-dimensional data, such as gene expression, where the joint complexity of anatomical and transcriptional patterns demands maximum compression. Established practice is to use standard principal component analysis (PCA), whose computational felicity is offset by limited expressivity, especially at great compression ratios. Employing whole-brain, voxel-wise Allen Brain Atlas transcription data, here we systematically compare compressed representations based on the most widely supported linear and non-linear methods-PCA, kernel PCA, non-negative matrix factorization (NMF), t-stochastic neighbour embedding (t-SNE), uniform manifold approximation and projection (UMAP), and deep auto-encoding-quantifying reconstruction fidelity, anatomical coherence, and predictive utility with respect to signalling, microstructural, and metabolic targets. We show that deep auto-encoders yield superior representations across all metrics of performance and target domains, supporting their use as the reference standard for representing transcription patterns in the human brain.
The legibility of the imaged human brain
Aug 23, 2023



Abstract:Our knowledge of the organisation of the human brain at the population-level is yet to translate into power to predict functional differences at the individual-level, limiting clinical applications, and casting doubt on the generalisability of inferred mechanisms. It remains unknown whether the difficulty arises from the absence of individuating biological patterns within the brain, or from limited power to access them with the models and compute at our disposal. Here we comprehensively investigate the resolvability of such patterns with data and compute at unprecedented scale. Across 23810 unique participants from UK Biobank, we systematically evaluate the predictability of 25 individual biological characteristics, from all available combinations of structural and functional neuroimaging data. Over 4526 GPU*hours of computation, we train, optimize, and evaluate out-of-sample 700 individual predictive models, including multilayer perceptrons of demographic, psychological, serological, chronic morbidity, and functional connectivity characteristics, and both uni- and multi-modal 3D convolutional neural network models of macro- and micro-structural brain imaging. We find a marked discrepancy between the high predictability of sex (balanced accuracy 99.7%), age (mean absolute error 2.048 years, R2 0.859), and weight (mean absolute error 2.609Kg, R2 0.625), for which we set new state-of-the-art performance, and the surprisingly low predictability of other characteristics. Neither structural nor functional imaging predicted individual psychology better than the coincidence of common chronic morbidity (p<0.05). Serology predicted common morbidity (p<0.05) and was best predicted by it (p<0.001), followed by structural neuroimaging (p<0.05). Our findings suggest either more informative imaging or more powerful models will be needed to decipher individual level characteristics from the brain.
The minimal computational substrate of fluid intelligence
Aug 14, 2023Abstract:The quantification of cognitive powers rests on identifying a behavioural task that depends on them. Such dependence cannot be assured, for the powers a task invokes cannot be experimentally controlled or constrained a priori, resulting in unknown vulnerability to failure of specificity and generalisability. Evaluating a compact version of Raven's Advanced Progressive Matrices (RAPM), a widely used clinical test of fluid intelligence, we show that LaMa, a self-supervised artificial neural network trained solely on the completion of partially masked images of natural environmental scenes, achieves human-level test scores a prima vista, without any task-specific inductive bias or training. Compared with cohorts of healthy and focally lesioned participants, LaMa exhibits human-like variation with item difficulty, and produces errors characteristic of right frontal lobe damage under degradation of its ability to integrate global spatial patterns. LaMa's narrow training and limited capacity -- comparable to the nervous system of the fruit fly -- suggest RAPM may be open to computationally simple solutions that need not necessarily invoke abstract reasoning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge