Nikolay Burlutskiy
Segment Anyword: Mask Prompt Inversion for Open-Set Grounded Segmentation
May 23, 2025Abstract:Open-set image segmentation poses a significant challenge because existing methods often demand extensive training or fine-tuning and generally struggle to segment unified objects consistently across diverse text reference expressions. Motivated by this, we propose Segment Anyword, a novel training-free visual concept prompt learning approach for open-set language grounded segmentation that relies on token-level cross-attention maps from a frozen diffusion model to produce segmentation surrogates or mask prompts, which are then refined into targeted object masks. Initial prompts typically lack coherence and consistency as the complexity of the image-text increases, resulting in suboptimal mask fragments. To tackle this issue, we further introduce a novel linguistic-guided visual prompt regularization that binds and clusters visual prompts based on sentence dependency and syntactic structural information, enabling the extraction of robust, noise-tolerant mask prompts, and significant improvements in segmentation accuracy. The proposed approach is effective, generalizes across different open-set segmentation tasks, and achieves state-of-the-art results of 52.5 (+6.8 relative) mIoU on Pascal Context 59, 67.73 (+25.73 relative) cIoU on gRefCOCO, and 67.4 (+1.1 relative to fine-tuned methods) mIoU on GranDf, which is the most complex open-set grounded segmentation task in the field.
MiceBoneChallenge: Micro-CT public dataset and six solutions for automatic growth plate detection in micro-CT mice bone scans
Nov 26, 2024



Abstract:Detecting and quantifying bone changes in micro-CT scans of rodents is a common task in preclinical drug development studies. However, this task is manual, time-consuming and subject to inter- and intra-observer variability. In 2024, Anonymous Company organized an internal challenge to develop models for automatic bone quantification. We prepared and annotated a high-quality dataset of 3D $\mu$CT bone scans from $83$ mice. The challenge attracted over $80$ AI scientists from around the globe who formed $23$ teams. The participants were tasked with developing a solution to identify the plane where the bone growth happens, which is essential for fully automatic segmentation of trabecular bone. As a result, six computer vision solutions were developed that can accurately identify the location of the growth plate plane. The solutions achieved the mean absolute error of $1.91\pm0.87$ planes from the ground truth on the test set, an accuracy level acceptable for practical use by a radiologist. The annotated 3D scans dataset along with the six solutions and source code, is being made public, providing researchers with opportunities to develop and benchmark their own approaches. The code, trained models, and the data will be shared.
Leveraging Transfer Learning and Multiple Instance Learning for HER2 Automatic Scoring of H\&E Whole Slide Images
Nov 05, 2024Abstract:Expression of human epidermal growth factor receptor 2 (HER2) is an important biomarker in breast cancer patients who can benefit from cost-effective automatic Hematoxylin and Eosin (H\&E) HER2 scoring. However, developing such scoring models requires large pixel-level annotated datasets. Transfer learning allows prior knowledge from different datasets to be reused while multiple-instance learning (MIL) allows the lack of detailed annotations to be mitigated. The aim of this work is to examine the potential of transfer learning on the performance of deep learning models pre-trained on (i) Immunohistochemistry (IHC) images, (ii) H\&E images and (iii) non-medical images. A MIL framework with an attention mechanism is developed using pre-trained models as patch-embedding models. It was found that embedding models pre-trained on H\&E images consistently outperformed the others, resulting in an average AUC-ROC value of $0.622$ across the 4 HER2 scores ($0.59-0.80$ per HER2 score). Furthermore, it was found that using multiple-instance learning with an attention layer not only allows for good classification results to be achieved, but it can also help with producing visual indication of HER2-positive areas in the H\&E slide image by utilising the patch-wise attention weights.
Lung tumor segmentation in MRI mice scans using 3D nnU-Net with minimum annotations
Nov 01, 2024



Abstract:In drug discovery, accurate lung tumor segmentation is an important step for assessing tumor size and its progression using \textit{in-vivo} imaging such as MRI. While deep learning models have been developed to automate this process, the focus has predominantly been on human subjects, neglecting the pivotal role of animal models in pre-clinical drug development. In this work, we focus on optimizing lung tumor segmentation in mice. First, we demonstrate that the nnU-Net model outperforms the U-Net, U-Net3+, and DeepMeta models. Most importantly, we achieve better results with nnU-Net 3D models than 2D models, indicating the importance of spatial context for segmentation tasks in MRI mice scans. This study demonstrates the importance of 3D input over 2D input images for lung tumor segmentation in MRI scans. Finally, we outperform the prior state-of-the-art approach that involves the combined segmentation of lungs and tumors within the lungs. Our work achieves comparable results using only lung tumor annotations requiring fewer annotations, saving time and annotation efforts. This work\footnote{\url{https://anonymous.4open.science/r/lung-tumour-mice-mri-64BB}} is an important step in automating pre-clinical animal studies to quantify the efficacy of experimental drugs, particularly in assessing tumor changes.
Can Medical Vision-Language Pre-training Succeed with Purely Synthetic Data?
Oct 17, 2024



Abstract:Medical Vision-Language Pre-training (MedVLP) has made significant progress in enabling zero-shot tasks for medical image understanding. However, training MedVLP models typically requires large-scale datasets with paired, high-quality image-text data, which are scarce in the medical domain. Recent advancements in Large Language Models (LLMs) and diffusion models have made it possible to generate large-scale synthetic image-text pairs. This raises the question: *Can MedVLP succeed using purely synthetic data?* To address this, we use off-the-shelf generative models to create synthetic radiology reports and paired Chest X-ray (CXR) images, and propose an automated pipeline to build a diverse, high-quality synthetic dataset, enabling a rigorous study that isolates model and training settings, focusing entirely from the data perspective. Our results show that MedVLP models trained *exclusively on synthetic data* outperform those trained on real data by **3.8%** in averaged AUC on zero-shot classification. Moreover, using a combination of synthetic and real data leads to a further improvement of **9.07%**. Additionally, MedVLP models trained on synthetic or mixed data consistently outperform those trained on real data in zero-shot grounding, as well as in fine-tuned classification and segmentation tasks. Our analysis suggests MedVLP trained on well-designed synthetic data can outperform models trained on real datasets, which may be limited by low-quality samples and long-tailed distributions.
Interpretable histopathology-based prediction of disease relevant features in Inflammatory Bowel Disease biopsies using weakly-supervised deep learning
Mar 20, 2023



Abstract:Crohn's Disease (CD) and Ulcerative Colitis (UC) are the two main Inflammatory Bowel Disease (IBD) types. We developed deep learning models to identify histological disease features for both CD and UC using only endoscopic labels. We explored fine-tuning and end-to-end training of two state-of-the-art self-supervised models for predicting three different endoscopic categories (i) CD vs UC (AUC=0.87), (ii) normal vs lesional (AUC=0.81), (iii) low vs high disease severity score (AUC=0.80). We produced visual attention maps to interpret what the models learned and validated them with the support of a pathologist, where we observed a strong association between the models' predictions and histopathological inflammatory features of the disease. Additionally, we identified several cases where the model incorrectly predicted normal samples as lesional but were correct on the microscopic level when reviewed by the pathologist. This tendency of histological presentation to be more severe than endoscopic presentation was previously published in the literature. In parallel, we utilised a model trained on the Colon Nuclei Identification and Counting (CoNIC) dataset to predict and explore 6 cell populations. We observed correlation between areas enriched with the predicted immune cells in biopsies and the pathologist's feedback on the attention maps. Finally, we identified several cell level features indicative of disease severity in CD and UC. These models can enhance our understanding about the pathology behind IBD and can shape our strategies for patient stratification in clinical trials.
Coupling Deep Imputation with Multitask Learning for Downstream Tasks on Genomics Data
Apr 28, 2022
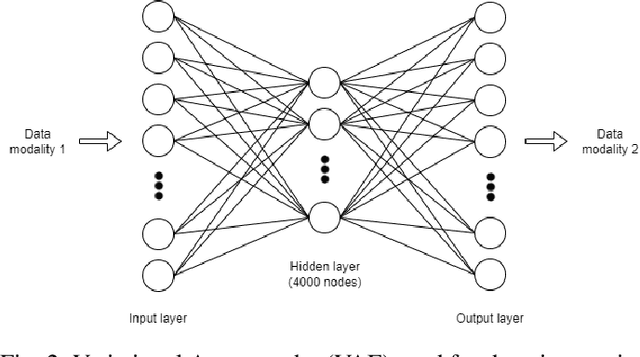
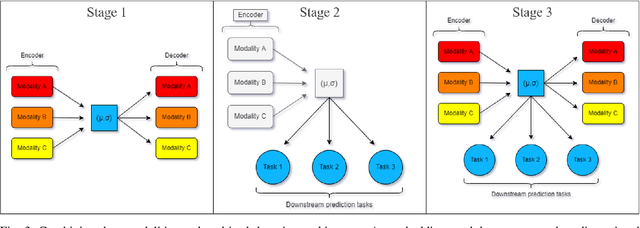
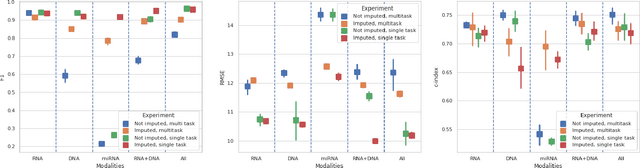
Abstract:Genomics data such as RNA gene expression, methylation and micro RNA expression are valuable sources of information for various clinical predictive tasks. For example, predicting survival outcomes, cancer histology type and other patients' related information is possible using not only clinical data but molecular data as well. Moreover, using these data sources together, for example in multitask learning, can boost the performance. However, in practice, there are many missing data points which leads to significantly lower patient numbers when analysing full cases, which in our setting refers to all modalities being present. In this paper we investigate how imputing data with missing values using deep learning coupled with multitask learning can help to reach state-of-the-art performance results using combined genomics modalities, RNA, micro RNA and methylation. We propose a generalised deep imputation method to impute values where a patient has all modalities present except one. Interestingly enough, deep imputation alone outperforms multitask learning alone for the classification and regression tasks across most combinations of modalities. In contrast, when using all modalities for survival prediction we observe that multitask learning alone outperforms deep imputation alone with statistical significance (adjusted p-value 0.03). Thus, both approaches are complementary when optimising performance for downstream predictive tasks.
Segmenting Potentially Cancerous Areas in Prostate Biopsies using Semi-Automatically Annotated Data
Apr 15, 2019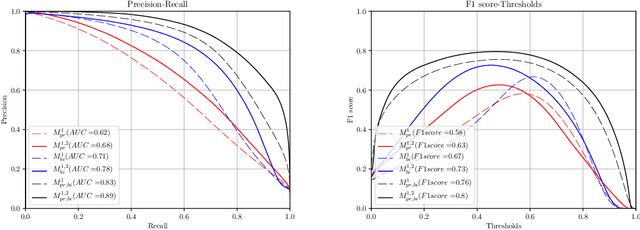
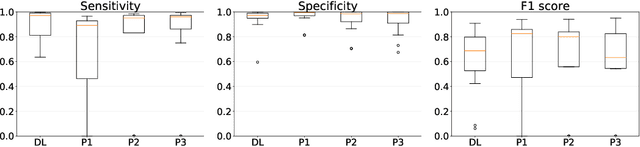
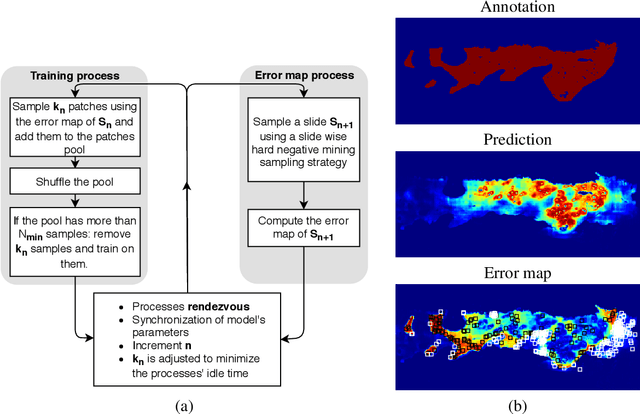

Abstract:Gleason grading specified in ISUP 2014 is the clinical standard in staging prostate cancer and the most important part of the treatment decision. However, the grading is subjective and suffers from high intra and inter-user variability. To improve the consistency and objectivity in the grading, we introduced glandular tissue WithOut Basal cells (WOB) as the ground truth. The presence of basal cells is the most accepted biomarker for benign glandular tissue and the absence of basal cells is a strong indicator of acinar prostatic adenocarcinoma, the most common form of prostate cancer. Glandular tissue can objectively be assessed as WOB or not WOB by using specific immunostaining for glandular tissue (Cytokeratin 8/18) and for basal cells (Cytokeratin 5/6 + p63). Even more, WOB allowed us to develop a semi-automated data generation pipeline to speed up the tremendously time consuming and expensive process of annotating whole slide images by pathologists. We generated 295 prostatectomy images exhaustively annotated with WOB. Then we used our Deep Learning Framework, which achieved the $2^{nd}$ best reported score in Camelyon17 Challenge, to train networks for segmenting WOB in needle biopsies. Evaluation of the model on 63 needle biopsies showed promising results which were improved further by finetuning the model on 118 biopsies annotated with WOB, achieving F1-score of 0.80 and Precision-Recall AUC of 0.89 at the pixel-level. Then we compared the performance of the model against 17 biopsies annotated independently by 3 pathologists using only H\&E staining. The comparison demonstrated that the model performed on a par with the pathologists. Finally, the model detected and accurately outlined existing WOB areas in two biopsies incorrectly annotated as totally WOB-free biopsies by three pathologists and in one biopsy by two pathologists.
A Deep Learning Framework for Automatic Diagnosis in Lung Cancer
Jul 27, 2018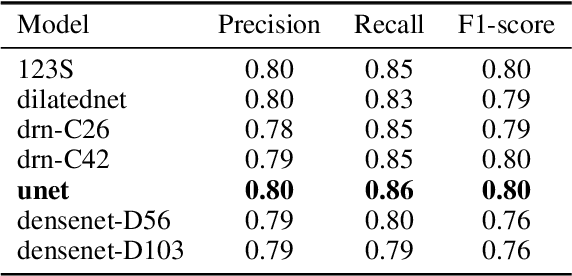
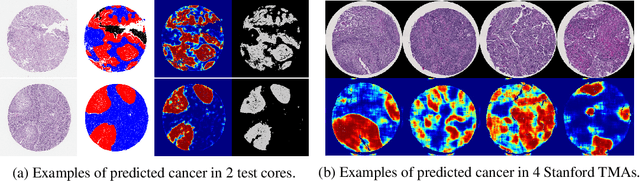
Abstract:We developed a deep learning framework that helps to automatically identify and segment lung cancer areas in patients' tissue specimens. The study was based on a cohort of lung cancer patients operated at the Uppsala University Hospital. The tissues were reviewed by lung pathologists and then the cores were compiled to tissue micro-arrays (TMAs). For experiments, hematoxylin-eosin stained slides from 712 patients were scanned and then manually annotated. Then these scans and annotations were used to train segmentation models of the developed framework. The performance of the developed deep learning framework was evaluated on fully annotated TMA cores from 178 patients reaching pixel-wise precision of 0.80 and recall of 0.86. Finally, publicly available Stanford TMA cores were used to demonstrate high performance of the framework qualitatively.
Multi-Resolution Networks for Semantic Segmentation in Whole Slide Images
Jul 25, 2018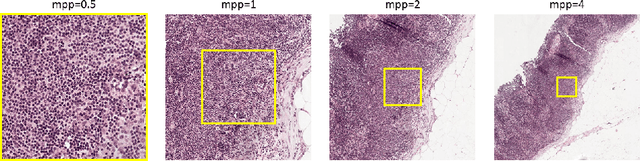
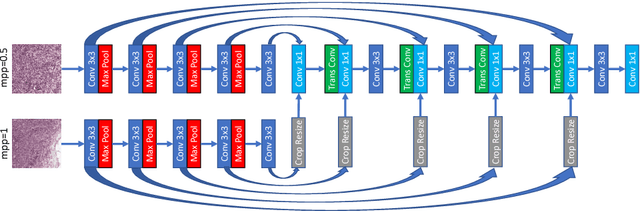
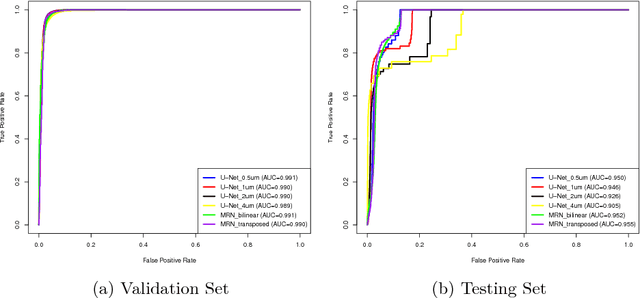
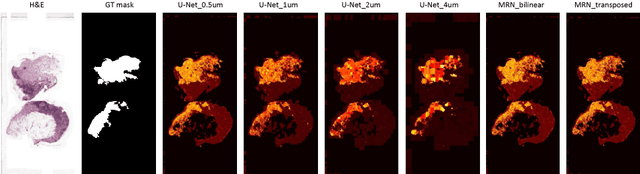
Abstract:Digital pathology provides an excellent opportunity for applying fully convolutional networks (FCNs) to tasks, such as semantic segmentation of whole slide images (WSIs). However, standard FCNs face challenges with respect to multi-resolution, inherited from the pyramid arrangement of WSIs. As a result, networks specifically designed to learn and aggregate information at different levels are desired. In this paper, we propose two novel multi-resolution networks based on the popular `U-Net' architecture, which are evaluated on a benchmark dataset for binary semantic segmentation in WSIs. The proposed methods outperform the U-Net, demonstrating superior learning and generalization capabilities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge