Nicolas Boutry
LRE
Improving segmentation of retinal arteries and veins using cardiac signal in doppler holograms
Nov 18, 2025Abstract:Doppler holography is an emerging retinal imaging technique that captures the dynamic behavior of blood flow with high temporal resolution, enabling quantitative assessment of retinal hemodynamics. This requires accurate segmentation of retinal arteries and veins, but traditional segmentation methods focus solely on spatial information and overlook the temporal richness of holographic data. In this work, we propose a simple yet effective approach for artery-vein segmentation in temporal Doppler holograms using standard segmentation architectures. By incorporating features derived from a dedicated pulse analysis pipeline, our method allows conventional U-Nets to exploit temporal dynamics and achieve performance comparable to more complex attention- or iteration-based models. These findings demonstrate that time-resolved preprocessing can unlock the full potential of deep learning for Doppler holography, opening new perspectives for quantitative exploration of retinal hemodynamics. The dataset is publicly available at https://huggingface.co/datasets/DigitalHolography/
GPML: Graph Processing for Machine Learning
May 13, 2025Abstract:The dramatic increase of complex, multi-step, and rapidly evolving attacks in dynamic networks involves advanced cyber-threat detectors. The GPML (Graph Processing for Machine Learning) library addresses this need by transforming raw network traffic traces into graph representations, enabling advanced insights into network behaviors. The library provides tools to detect anomalies in interaction and community shifts in dynamic networks. GPML supports community and spectral metrics extraction, enhancing both real-time detection and historical forensics analysis. This library supports modern cybersecurity challenges with a robust, graph-based approach.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
Reasoning with trees: interpreting CNNs using hierarchies
Jun 19, 2024Abstract:Challenges persist in providing interpretable explanations for neural network reasoning in explainable AI (xAI). Existing methods like Integrated Gradients produce noisy maps, and LIME, while intuitive, may deviate from the model's reasoning. We introduce a framework that uses hierarchical segmentation techniques for faithful and interpretable explanations of Convolutional Neural Networks (CNNs). Our method constructs model-based hierarchical segmentations that maintain the model's reasoning fidelity and allows both human-centric and model-centric segmentation. This approach offers multiscale explanations, aiding bias identification and enhancing understanding of neural network decision-making. Experiments show that our framework, xAiTrees, delivers highly interpretable and faithful model explanations, not only surpassing traditional xAI methods but shedding new light on a novel approach to enhancing xAI interpretability. Code at: https://github.com/CarolMazini/reasoning_with_trees .
Transforming gradient-based techniques into interpretable methods
Jan 25, 2024



Abstract:The explication of Convolutional Neural Networks (CNN) through xAI techniques often poses challenges in interpretation. The inherent complexity of input features, notably pixels extracted from images, engenders complex correlations. Gradient-based methodologies, exemplified by Integrated Gradients (IG), effectively demonstrate the significance of these features. Nevertheless, the conversion of these explanations into images frequently yields considerable noise. Presently, we introduce GAD (Gradient Artificial Distancing) as a supportive framework for gradient-based techniques. Its primary objective is to accentuate influential regions by establishing distinctions between classes. The essence of GAD is to limit the scope of analysis during visualization and, consequently reduce image noise. Empirical investigations involving occluded images have demonstrated that the identified regions through this methodology indeed play a pivotal role in facilitating class differentiation.
Unsupervised discovery of Interpretable Visual Concepts
Aug 31, 2023Abstract:Providing interpretability of deep-learning models to non-experts, while fundamental for a responsible real-world usage, is challenging. Attribution maps from xAI techniques, such as Integrated Gradients, are a typical example of a visualization technique containing a high level of information, but with difficult interpretation. In this paper, we propose two methods, Maximum Activation Groups Extraction (MAGE) and Multiscale Interpretable Visualization (Ms-IV), to explain the model's decision, enhancing global interpretability. MAGE finds, for a given CNN, combinations of features which, globally, form a semantic meaning, that we call concepts. We group these similar feature patterns by clustering in ``concepts'', that we visualize through Ms-IV. This last method is inspired by Occlusion and Sensitivity analysis (incorporating causality), and uses a novel metric, called Class-aware Order Correlation (CaOC), to globally evaluate the most important image regions according to the model's decision space. We compare our approach to xAI methods such as LIME and Integrated Gradients. Experimental results evince the Ms-IV higher localization and faithfulness values. Finally, qualitative evaluation of combined MAGE and Ms-IV demonstrate humans' ability to agree, based on the visualization, on the decision of clusters' concepts; and, to detect, among a given set of networks, the existence of bias.
BuyTheDips: PathLoss for improved topology-preserving deep learning-based image segmentation
Jul 23, 2022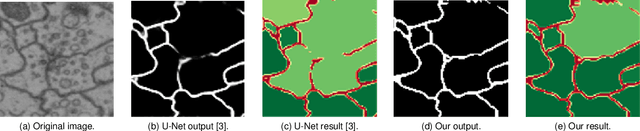
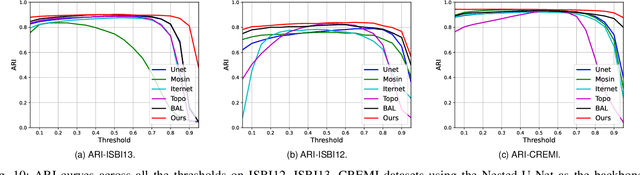
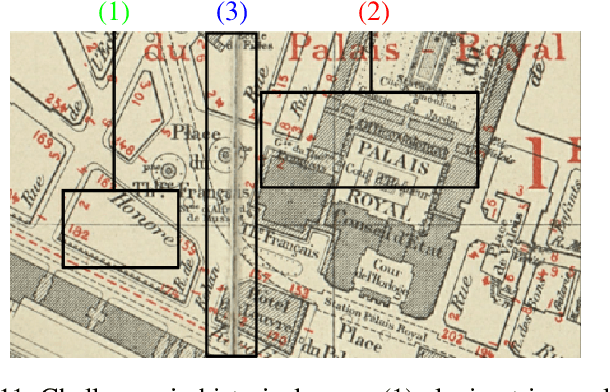
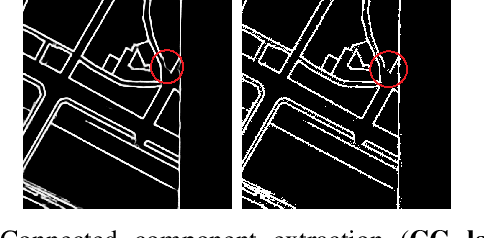
Abstract:Capturing the global topology of an image is essential for proposing an accurate segmentation of its domain. However, most of existing segmentation methods do not preserve the initial topology of the given input, which is detrimental for numerous downstream object-based tasks. This is all the more true for deep learning models which most work at local scales. In this paper, we propose a new topology-preserving deep image segmentation method which relies on a new leakage loss: the Pathloss. Our method is an extension of the BALoss [1], in which we want to improve the leakage detection for better recovering the closeness property of the image segmentation. This loss allows us to correctly localize and fix the critical points (a leakage in the boundaries) that could occur in the predictions, and is based on a shortest-path search algorithm. This way, loss minimization enforces connectivity only where it is necessary and finally provides a good localization of the boundaries of the objects in the image. Moreover, according to our research, our Pathloss learns to preserve stronger elongated structure compared to methods without using topology-preserving loss. Training with our topological loss function, our method outperforms state-of-the-art topology-aware methods on two representative datasets of different natures: Electron Microscopy and Historical Map.
A Proof of the Tree of Shapes in n-D
Jun 10, 2022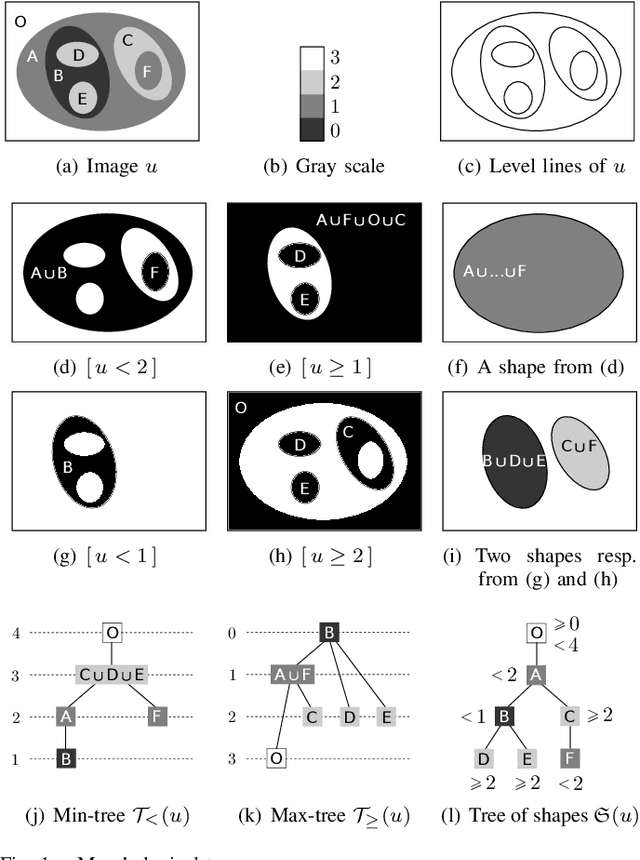
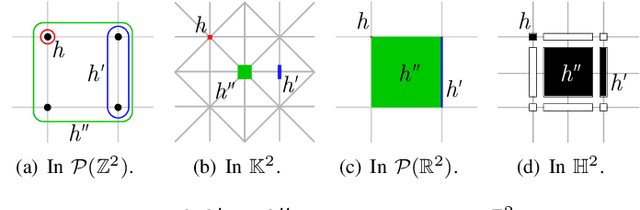
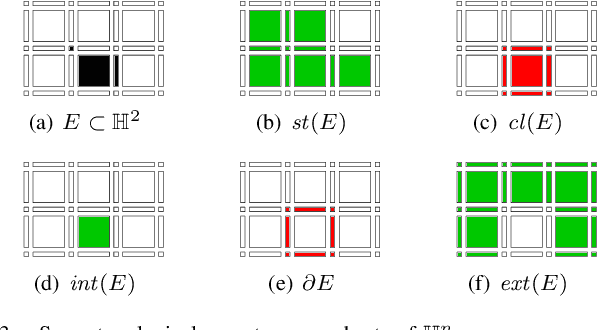
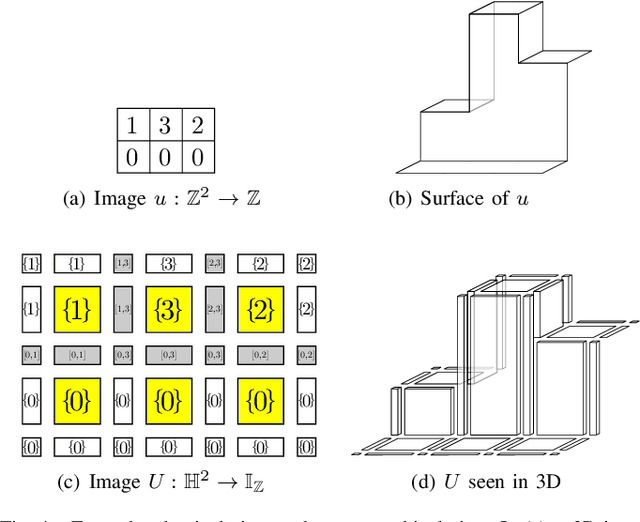
Abstract:In this paper, we prove that the self-dual morphological hierarchical structure computed on a n-D gray-level wellcomposed image u by the algorithm of G{\'e}raud et al. [1] is exactly the mathematical structure defined to be the tree of shape of u in Najman et al [2]. We recall that this algorithm is in quasi-linear time and thus considered to be optimal. The tree of shapes leads to many applications in mathematical morphology and in image processing like grain filtering, shapings, image segmentation, and so on.
Some equivalence relation between persistent homology and morphological dynamics
May 25, 2022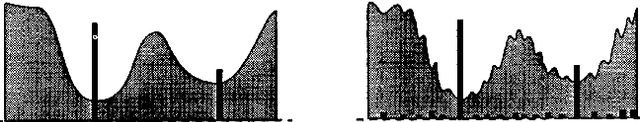
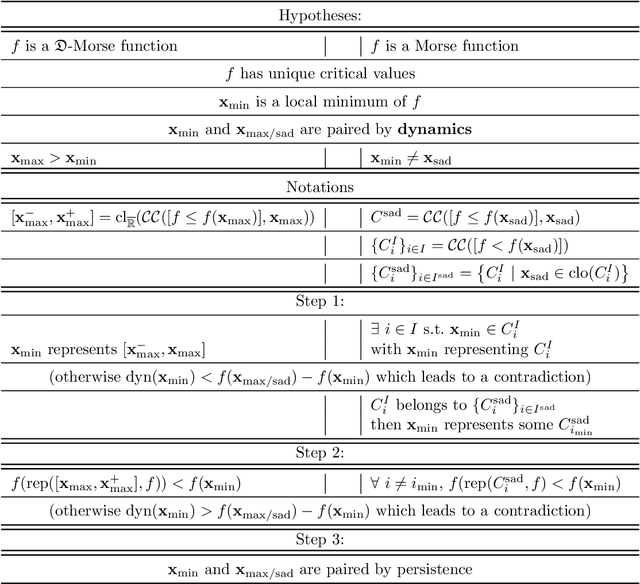
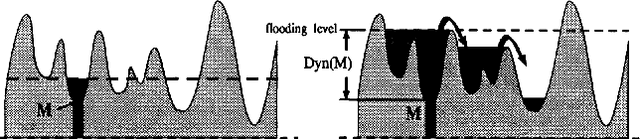
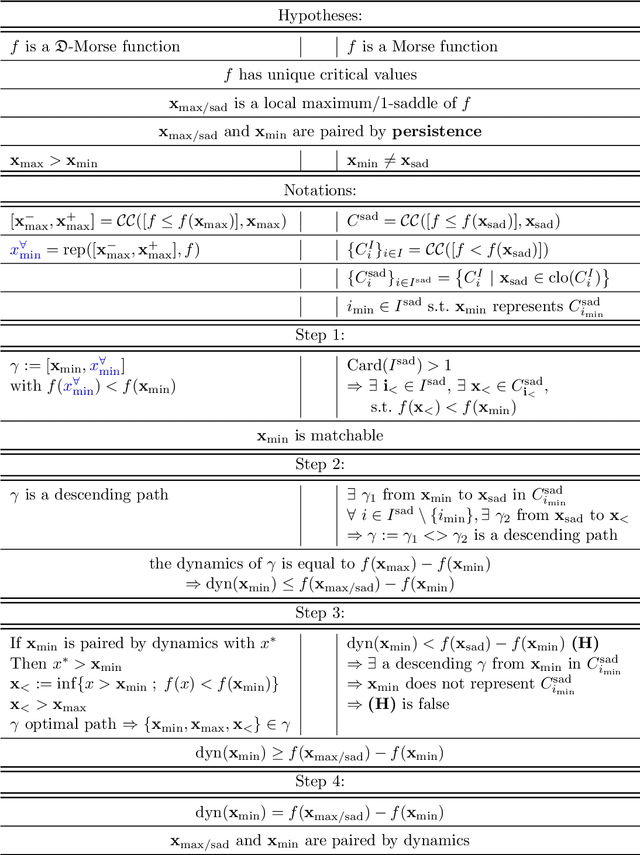
Abstract:In Mathematical Morphology (MM), connected filters based on dynamics are used to filter the extrema of an image. Similarly, persistence is a concept coming from Persistent Homology (PH) and Morse Theory (MT) that represents the stability of the extrema of a Morse function. Since these two concepts seem to be closely related, in this paper we examine their relationship, and we prove that they are equal on n-D Morse functions, n $\ge$ 1. More exactly, pairing a minimum with a 1-saddle by dynamics or pairing the same 1-saddle with a minimum by persistence leads exactly to the same pairing, assuming that the critical values of the studied Morse function are unique. This result is a step further to show how much topological data analysis and mathematical morphology are related, paving the way for a more in-depth study of the relations between these two research fields.
Local Intensity Order Transformation for Robust Curvilinear Object Segmentation
Feb 25, 2022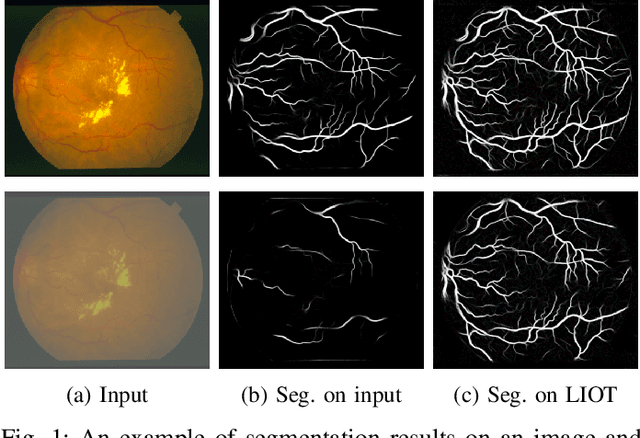
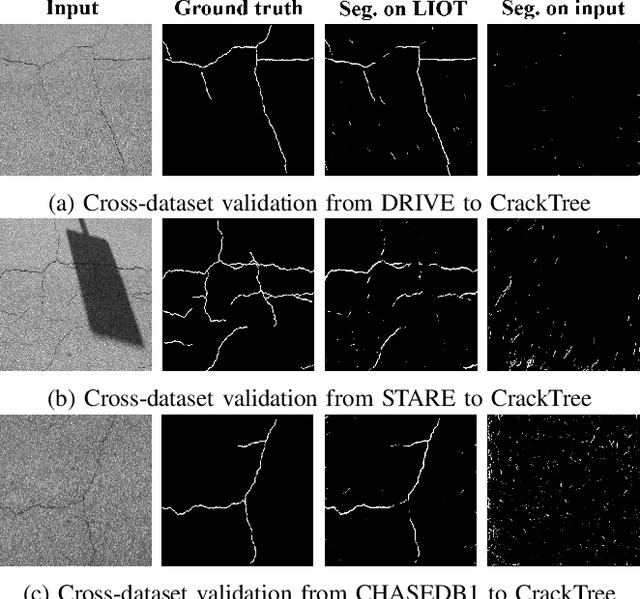
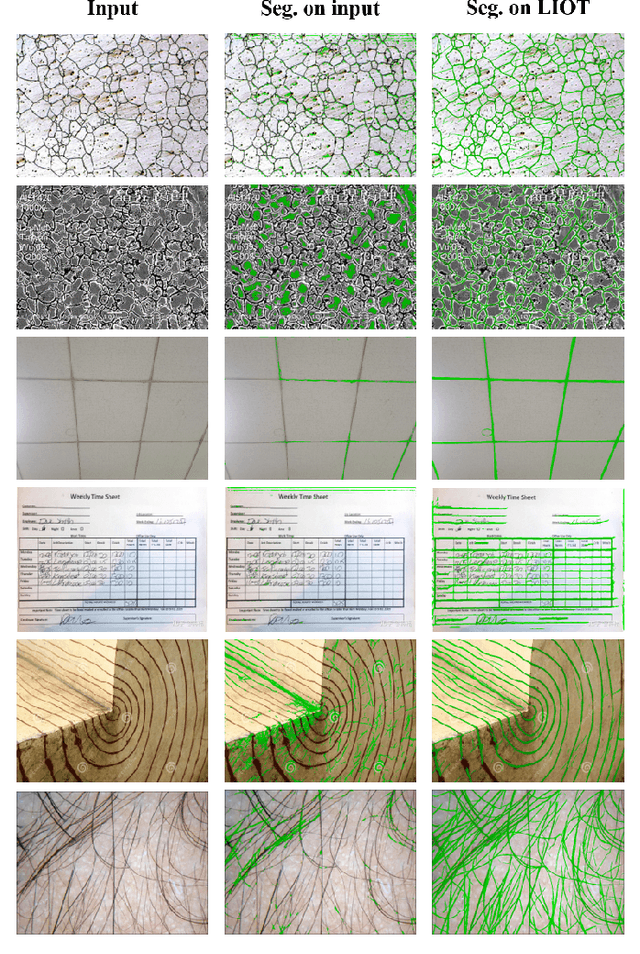
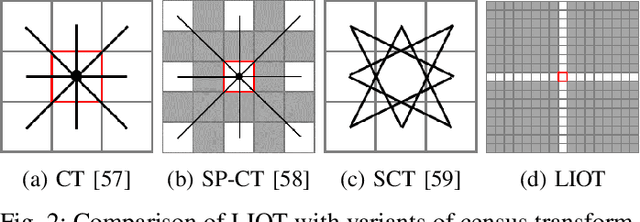
Abstract:Segmentation of curvilinear structures is important in many applications, such as retinal blood vessel segmentation for early detection of vessel diseases and pavement crack segmentation for road condition evaluation and maintenance. Currently, deep learning-based methods have achieved impressive performance on these tasks. Yet, most of them mainly focus on finding powerful deep architectures but ignore capturing the inherent curvilinear structure feature (e.g., the curvilinear structure is darker than the context) for a more robust representation. In consequence, the performance usually drops a lot on cross-datasets, which poses great challenges in practice. In this paper, we aim to improve the generalizability by introducing a novel local intensity order transformation (LIOT). Specifically, we transfer a gray-scale image into a contrast-invariant four-channel image based on the intensity order between each pixel and its nearby pixels along with the four (horizontal and vertical) directions. This results in a representation that preserves the inherent characteristic of the curvilinear structure while being robust to contrast changes. Cross-dataset evaluation on three retinal blood vessel segmentation datasets demonstrates that LIOT improves the generalizability of some state-of-the-art methods. Additionally, the cross-dataset evaluation between retinal blood vessel segmentation and pavement crack segmentation shows that LIOT is able to preserve the inherent characteristic of curvilinear structure with large appearance gaps. An implementation of the proposed method is available at https://github.com/TY-Shi/LIOT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge