Muhammad Asad
Spatial gradient consistency for unsupervised learning of hyperspectral demosaicking: Application to surgical imaging
Feb 21, 2023Abstract:Hyperspectral imaging has the potential to improve intraoperative decision making if tissue characterisation is performed in real-time and with high-resolution. Hyperspectral snapshot mosaic sensors offer a promising approach due to their fast acquisition speed and compact size. However, a demosaicking algorithm is required to fully recover the spatial and spectral information of the snapshot images. Most state-of-the-art demosaicking algorithms require ground-truth training data with paired snapshot and high-resolution hyperspectral images, but such imagery pairs with the exact same scene are physically impossible to acquire in intraoperative settings. In this work, we present a fully unsupervised hyperspectral image demosaicking algorithm which only requires exemplar snapshot images for training purposes. We regard hyperspectral demosaicking as an ill-posed linear inverse problem which we solve using a deep neural network. We take advantage of the spectral correlation occurring in natural scenes to design a novel inter spectral band regularisation term based on spatial gradient consistency. By combining our proposed term with standard regularisation techniques and exploiting a standard data fidelity term, we obtain an unsupervised loss function for training deep neural networks, which allows us to achieve real-time hyperspectral image demosaicking. Quantitative results on hyperspetral image datasets show that our unsupervised demosaicking approach can achieve similar performance to its supervised counter-part, and significantly outperform linear demosaicking. A qualitative user study on real snapshot hyperspectral surgical images confirms the results from the quantitative analysis. Our results suggest that the proposed unsupervised algorithm can achieve promising hyperspectral demosaicking in real-time thus advancing the suitability of the modality for intraoperative use.
Boundary Distance Loss for Intra-/Extra-meatal Segmentation of Vestibular Schwannoma
Aug 09, 2022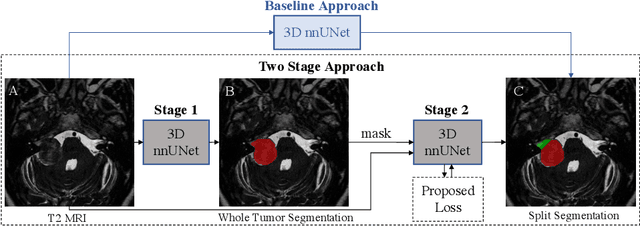
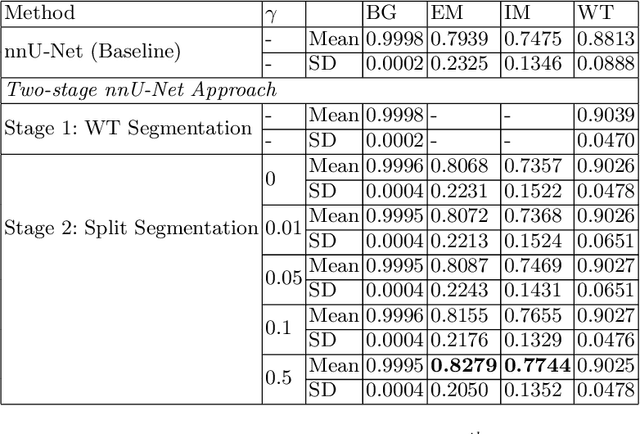
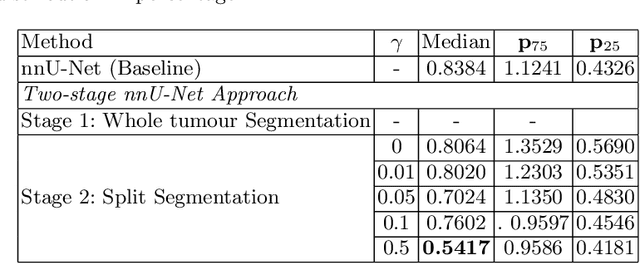
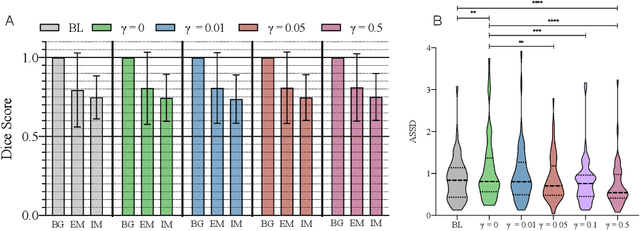
Abstract:Vestibular Schwannoma (VS) typically grows from the inner ear to the brain. It can be separated into two regions, intrameatal and extrameatal respectively corresponding to being inside or outside the inner ear canal. The growth of the extrameatal regions is a key factor that determines the disease management followed by the clinicians. In this work, a VS segmentation approach with subdivision into intra-/extra-meatal parts is presented. We annotated a dataset consisting of 227 T2 MRI instances, acquired longitudinally on 137 patients, excluding post-operative instances. We propose a staged approach, with the first stage performing the whole tumour segmentation and the second stage performing the intra-/extra-meatal segmentation using the T2 MRI along with the mask obtained from the first stage. To improve on the accuracy of the predicted meatal boundary, we introduce a task-specific loss which we call Boundary Distance Loss. The performance is evaluated in contrast to the direct intrameatal extrameatal segmentation task performance, i.e. the Baseline. Our proposed method, with the two-stage approach and the Boundary Distance Loss, achieved a Dice score of 0.8279+-0.2050 and 0.7744+-0.1352 for extrameatal and intrameatal regions respectively, significantly improving over the Baseline, which gave Dice score of 0.7939+-0.2325 and 0.7475+-0.1346 for the extrameatal and intrameatal regions respectively.
FastGeodis: Fast Generalised Geodesic Distance Transform
Jul 26, 2022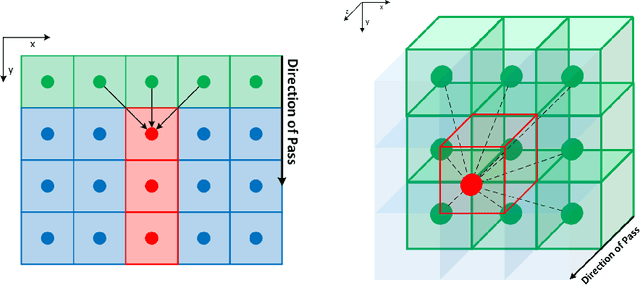
Abstract:The FastGeodis package provides an efficient implementation for computing Geodesic and Euclidean distance transforms (or a mixture of both) targeting efficient utilisation of CPU and GPU hardwares. In particular, it implements paralellisable raster scan method from Criminisi et al, where elements in row (2D) or plane (3D) can be computed with parallel threads. This package is able to handle 2D as well as 3D data where it achieves up to 15x speed-up on CPU and up to 60x speed-up on GPU as compared to existing open-source libraries, which uses non-parallelisable single-thread CPU implementation. The performance speed-ups reported here were evaluated using 3D volume data on Nvidia GeForce Titan X (12 GB) with 6-Core Intel Xeon E5-1650 CPU. This package is available at: https://github.com/masadcv/FastGeodis
MONAI Label: A framework for AI-assisted Interactive Labeling of 3D Medical Images
Mar 23, 2022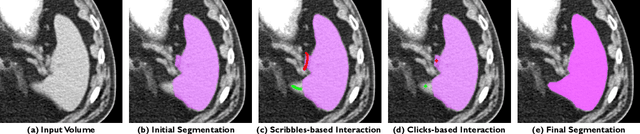
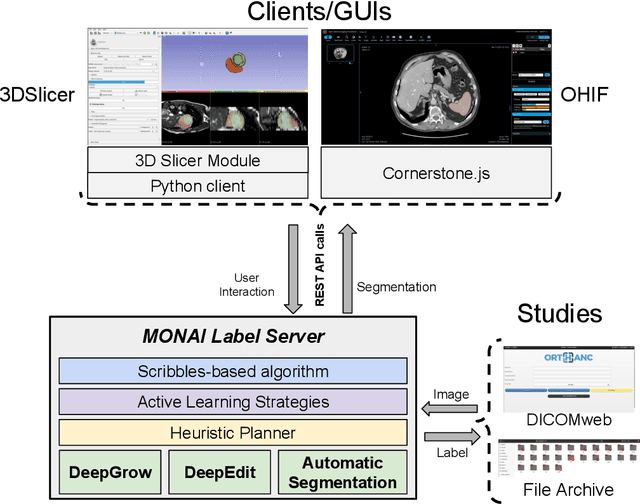
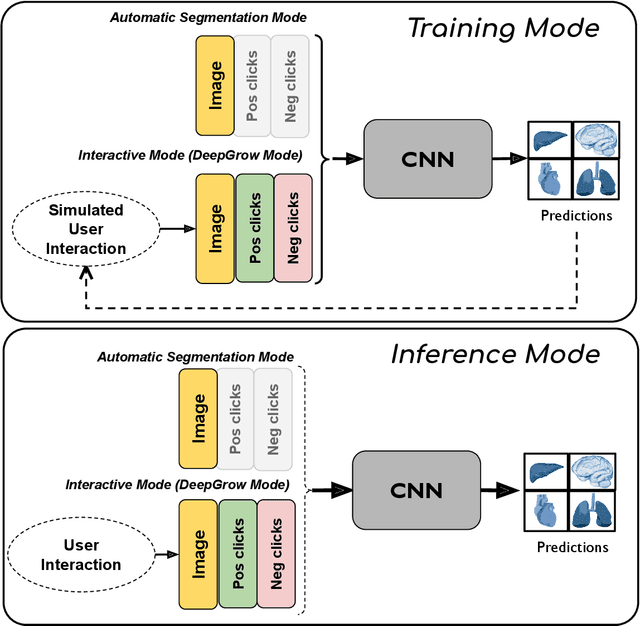
Abstract:The lack of annotated datasets is a major challenge in training new task-specific supervised AI algorithms as manual annotation is expensive and time-consuming. To address this problem, we present MONAI Label, a free and open-source platform that facilitates the development of AI-based applications that aim at reducing the time required to annotate 3D medical image datasets. Through MONAI Label researchers can develop annotation applications focusing on their domain of expertise. It allows researchers to readily deploy their apps as services, which can be made available to clinicians via their preferred user-interface. Currently, MONAI Label readily supports locally installed (3DSlicer) and web-based (OHIF) frontends, and offers two Active learning strategies to facilitate and speed up the training of segmentation algorithms. MONAI Label allows researchers to make incremental improvements to their labeling apps by making them available to other researchers and clinicians alike. Lastly, MONAI Label provides sample labeling apps, namely DeepEdit and DeepGrow, demonstrating dramatically reduced annotation times.
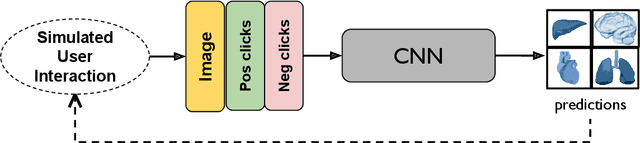
ECONet: Efficient Convolutional Online Likelihood Network for Scribble-based Interactive Segmentation
Feb 08, 2022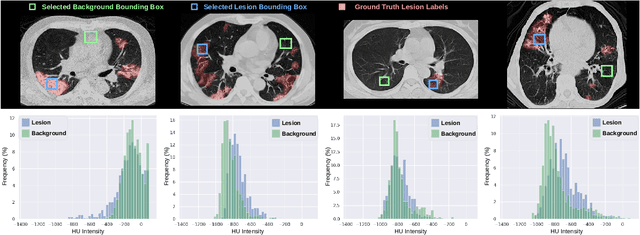

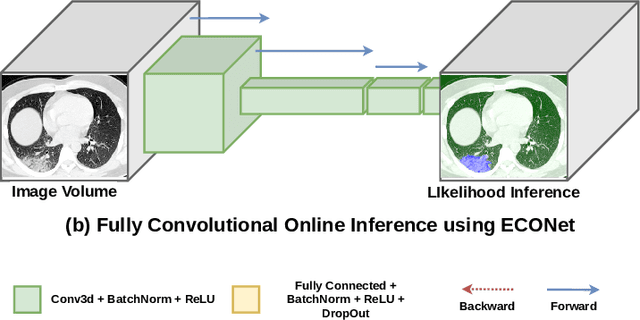
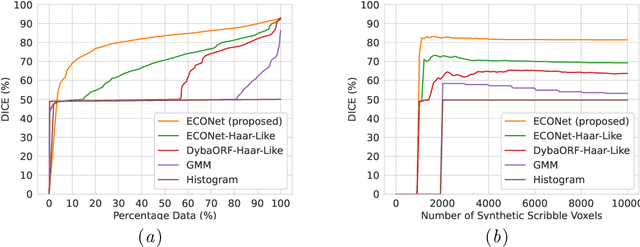
Abstract:Automatic segmentation of lung lesions associated with COVID-19 in CT images requires large amount of annotated volumes. Annotations mandate expert knowledge and are time-intensive to obtain through fully manual segmentation methods. Additionally, lung lesions have large inter-patient variations, with some pathologies having similar visual appearance as healthy lung tissues. This poses a challenge when applying existing semi-automatic interactive segmentation techniques for data labelling. To address these challenges, we propose an efficient convolutional neural networks (CNNs) that can be learned online while the annotator provides scribble-based interaction. To accelerate learning from only the samples labelled through user-interactions, a patch-based approach is used for training the network. Moreover, we use weighted cross-entropy loss to address the class imbalance that may result from user-interactions. During online inference, the learned network is applied to the whole input volume using a fully convolutional approach. We compare our proposed method with state-of-the-art using synthetic scribbles and show that it outperforms existing methods on the task of annotating lung lesions associated with COVID-19, achieving 16% higher Dice score while reducing execution time by 3$\times$ and requiring 9000 lesser scribbles-based labelled voxels. Due to the online learning aspect, our approach adapts quickly to user input, resulting in high quality segmentation labels. Source code for ECONet is available at: https://github.com/masadcv/ECONet-MONAILabel
Federated Learning Versus Classical Machine Learning: A Convergence Comparison
Jul 22, 2021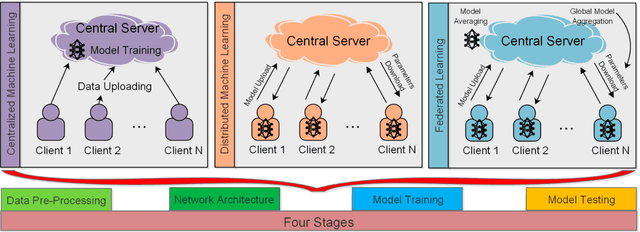
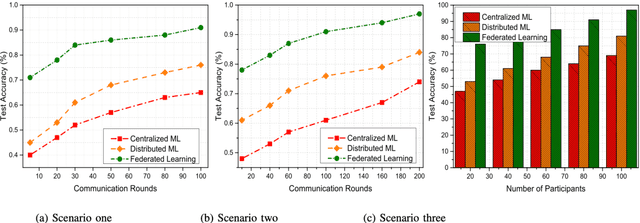
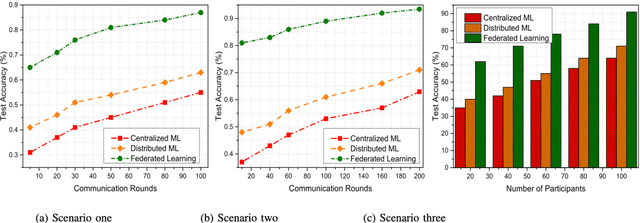
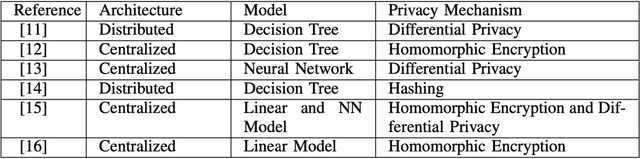
Abstract:In the past few decades, machine learning has revolutionized data processing for large scale applications. Simultaneously, increasing privacy threats in trending applications led to the redesign of classical data training models. In particular, classical machine learning involves centralized data training, where the data is gathered, and the entire training process executes at the central server. Despite significant convergence, this training involves several privacy threats on participants' data when shared with the central cloud server. To this end, federated learning has achieved significant importance over distributed data training. In particular, the federated learning allows participants to collaboratively train the local models on local data without revealing their sensitive information to the central cloud server. In this paper, we perform a convergence comparison between classical machine learning and federated learning on two publicly available datasets, namely, logistic-regression-MNIST dataset and image-classification-CIFAR-10 dataset. The simulation results demonstrate that federated learning achieves higher convergence within limited communication rounds while maintaining participants' anonymity. We hope that this research will show the benefits and help federated learning to be implemented widely.
Evaluating the Communication Efficiency in Federated Learning Algorithms
Apr 06, 2020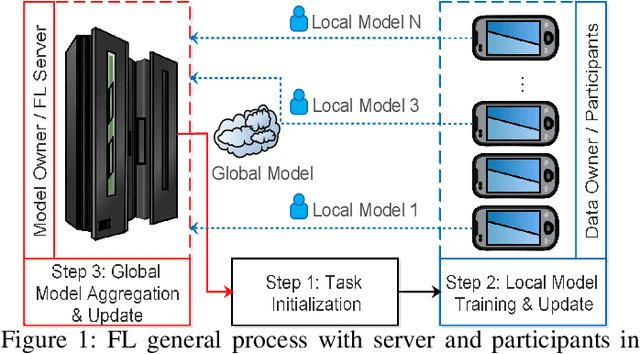
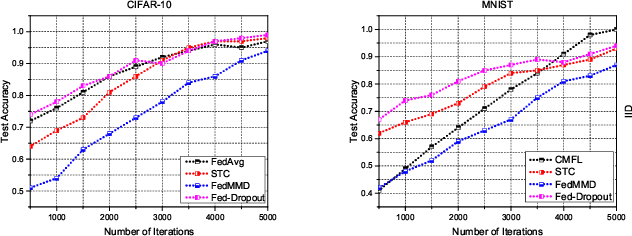
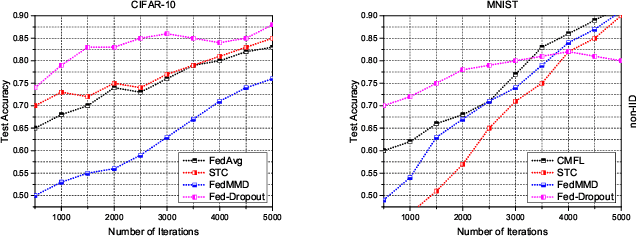
Abstract:In the era of advanced technologies, mobile devices are equipped with computing and sensing capabilities that gather excessive amounts of data. These amounts of data are suitable for training different learning models. Cooperated with advancements in Deep Learning (DL), these learning models empower numerous useful applications, e.g., image processing, speech recognition, healthcare, vehicular network and many more. Traditionally, Machine Learning (ML) approaches require data to be centralised in cloud-based data-centres. However, this data is often large in quantity and privacy-sensitive which prevents logging into these data-centres for training the learning models. In turn, this results in critical issues of high latency and communication inefficiency. Recently, in light of new privacy legislations in many countries, the concept of Federated Learning (FL) has been introduced. In FL, mobile users are empowered to learn a global model by aggregating their local models, without sharing the privacy-sensitive data. Usually, these mobile users have slow network connections to the data-centre where the global model is maintained. Moreover, in a complex and large scale network, heterogeneous devices that have various energy constraints are involved. This raises the challenge of communication cost when implementing FL at large scale. To this end, in this research, we begin with the fundamentals of FL, and then, we highlight the recent FL algorithms and evaluate their communication efficiency with detailed comparisons. Furthermore, we propose a set of solutions to alleviate the existing FL problems both from communication perspective and privacy perspective.
PROPEL: Probabilistic Parametric Regression Loss for Convolutional Neural Networks
Jul 28, 2018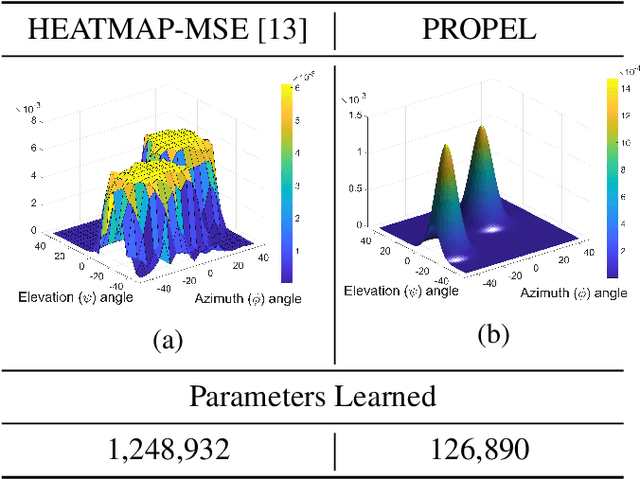
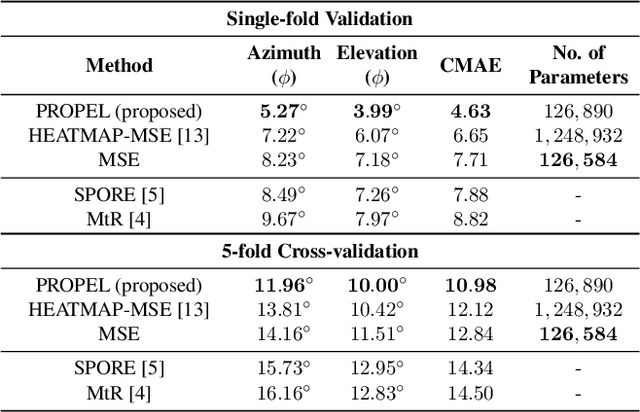
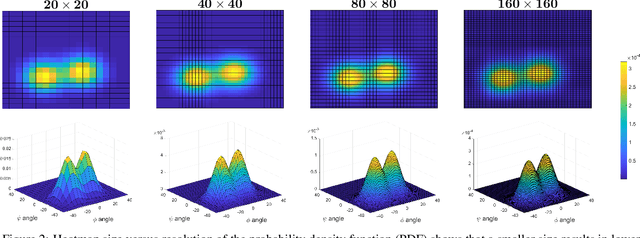
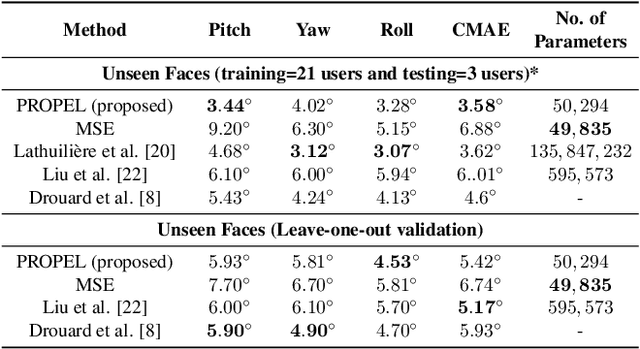
Abstract:Recently, Convolutional Neural Networks (CNNs) have dominated the field of computer vision. Their widespread success has been attributed to their representation learning capabilities. For classification tasks, CNNs have widely employed probabilistic output and have shown the significance of providing additional confidence for predictions. However, such probabilistic methodologies are not widely applicable for addressing regression problems using CNNs, as regression involves learning unconstrained continuous and, in many cases, multi-variate target variables. We propose a PRObabilistic Parametric rEgression Loss (PROPEL) that enables probabilistic regression using CNNs. PROPEL is fully differentiable and, hence, can be easily incorporated for end-to-end training of existing regressive CNN architectures. The proposed method is flexible as it learns complex unconstrained probabilities while being generalizable to higher dimensional multi-variate regression problems. We utilize a PROPEL-based CNN to address the problem of learning hand and head orientation from uncalibrated color images. Comprehensive experimental validation and comparisons with existing CNN regression loss functions are provided. Our experimental results indicate that PROPEL significantly improves the performance of a CNN, while reducing model parameters by 10x as compared to the existing state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge