Mina Rezaei
Joint Debiased Representation and Image Clustering Learning with Self-Supervision
Sep 14, 2022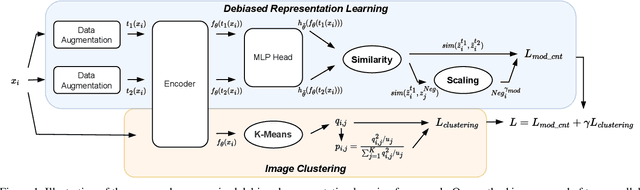
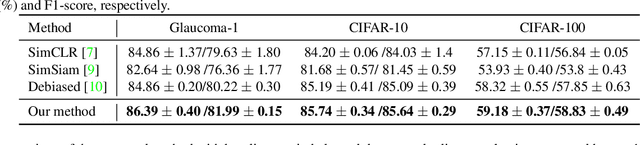
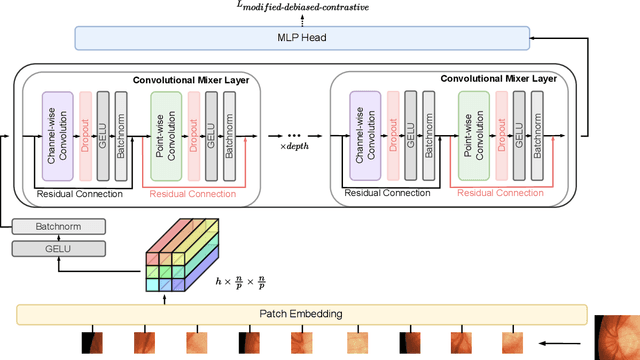
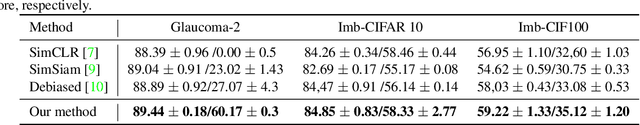
Abstract:Contrastive learning is among the most successful methods for visual representation learning, and its performance can be further improved by jointly performing clustering on the learned representations. However, existing methods for joint clustering and contrastive learning do not perform well on long-tailed data distributions, as majority classes overwhelm and distort the loss of minority classes, thus preventing meaningful representations to be learned. Motivated by this, we develop a novel joint clustering and contrastive learning framework by adapting the debiased contrastive loss to avoid under-clustering minority classes of imbalanced datasets. We show that our proposed modified debiased contrastive loss and divergence clustering loss improves the performance across multiple datasets and learning tasks. The source code is available at https://anonymous.4open.science/r/SSL-debiased-clustering
Robust and Efficient Imbalanced Positive-Unlabeled Learning with Self-supervision
Sep 06, 2022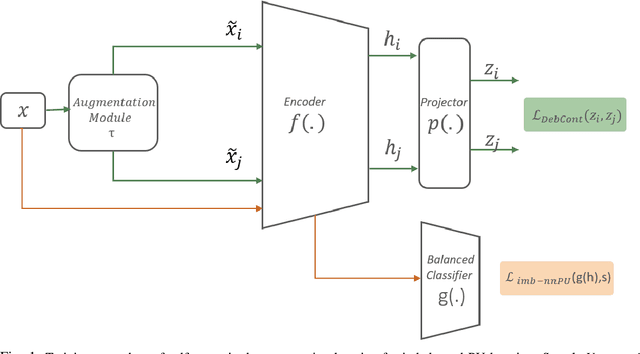
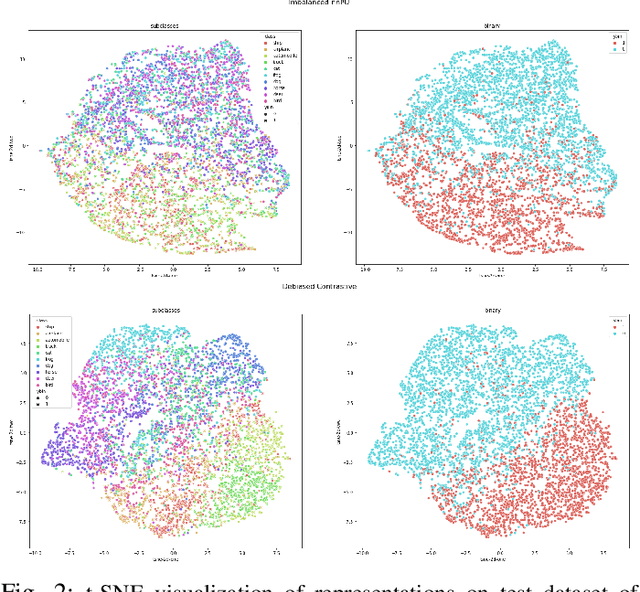
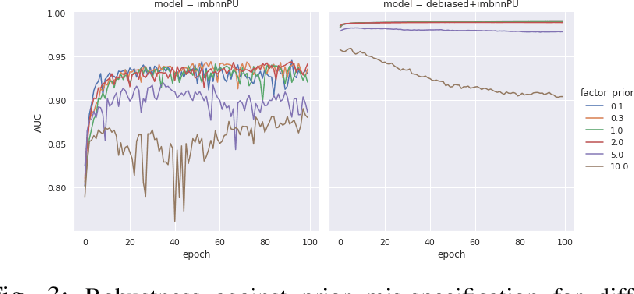
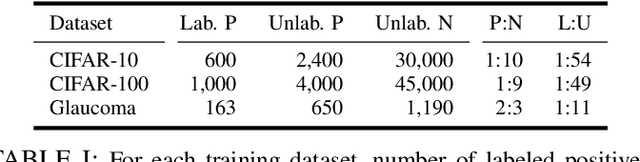
Abstract:Learning from positive and unlabeled (PU) data is a setting where the learner only has access to positive and unlabeled samples while having no information on negative examples. Such PU setting is of great importance in various tasks such as medical diagnosis, social network analysis, financial markets analysis, and knowledge base completion, which also tend to be intrinsically imbalanced, i.e., where most examples are actually negatives. Most existing approaches for PU learning, however, only consider artificially balanced datasets and it is unclear how well they perform in the realistic scenario of imbalanced and long-tail data distribution. This paper proposes to tackle this challenge via robust and efficient self-supervised pretraining. However, training conventional self-supervised learning methods when applied with highly imbalanced PU distribution needs better reformulation. In this paper, we present \textit{ImPULSeS}, a unified representation learning framework for \underline{Im}balanced \underline{P}ositive \underline{U}nlabeled \underline{L}earning leveraging \underline{Se}lf-\underline{S}upervised debiase pre-training. ImPULSeS uses a generic combination of large-scale unsupervised learning with debiased contrastive loss and additional reweighted PU loss. We performed different experiments across multiple datasets to show that ImPULSeS is able to halve the error rate of the previous state-of-the-art, even compared with previous methods that are given the true prior. Moreover, our method showed increased robustness to prior misspecification and superior performance even when pretraining was performed on an unrelated dataset. We anticipate such robustness and efficiency will make it much easier for practitioners to obtain excellent results on other PU datasets of interest. The source code is available at \url{https://github.com/JSchweisthal/ImPULSeS}
FiLM-Ensemble: Probabilistic Deep Learning via Feature-wise Linear Modulation
May 31, 2022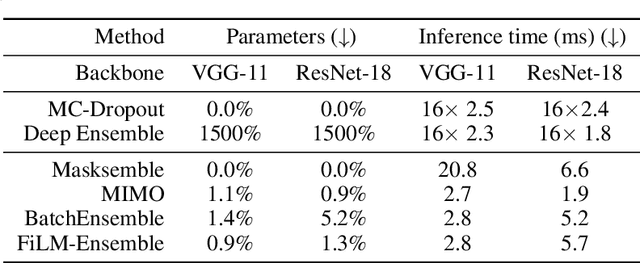
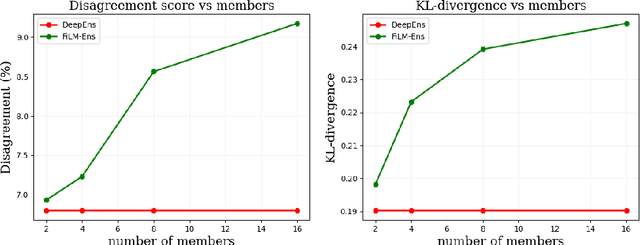
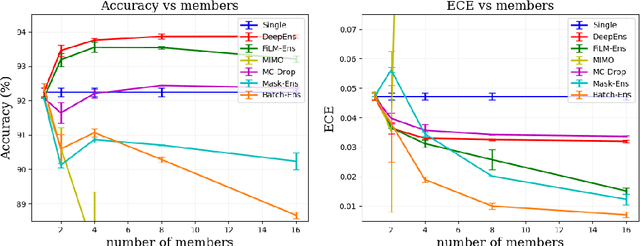
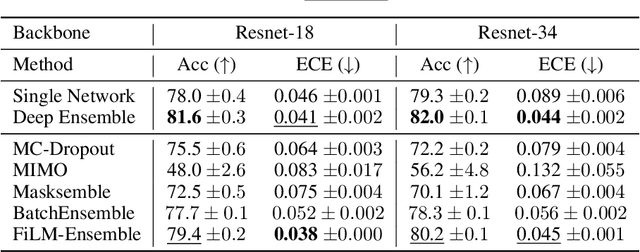
Abstract:The ability to estimate epistemic uncertainty is often crucial when deploying machine learning in the real world, but modern methods often produce overconfident, uncalibrated uncertainty predictions. A common approach to quantify epistemic uncertainty, usable across a wide class of prediction models, is to train a model ensemble. In a naive implementation, the ensemble approach has high computational cost and high memory demand. This challenges in particular modern deep learning, where even a single deep network is already demanding in terms of compute and memory, and has given rise to a number of attempts to emulate the model ensemble without actually instantiating separate ensemble members. We introduce FiLM-Ensemble, a deep, implicit ensemble method based on the concept of Feature-wise Linear Modulation (FiLM). That technique was originally developed for multi-task learning, with the aim of decoupling different tasks. We show that the idea can be extended to uncertainty quantification: by modulating the network activations of a single deep network with FiLM, one obtains a model ensemble with high diversity, and consequently well-calibrated estimates of epistemic uncertainty, with low computational overhead in comparison. Empirically, FiLM-Ensemble outperforms other implicit ensemble methods, and it and comes very close to the upper bound of an explicit ensemble of networks (sometimes even beating it), at a fraction of the memory cost.
Analyzing the Effects of Handling Data Imbalance on Learned Features from Medical Images by Looking Into the Models
Apr 04, 2022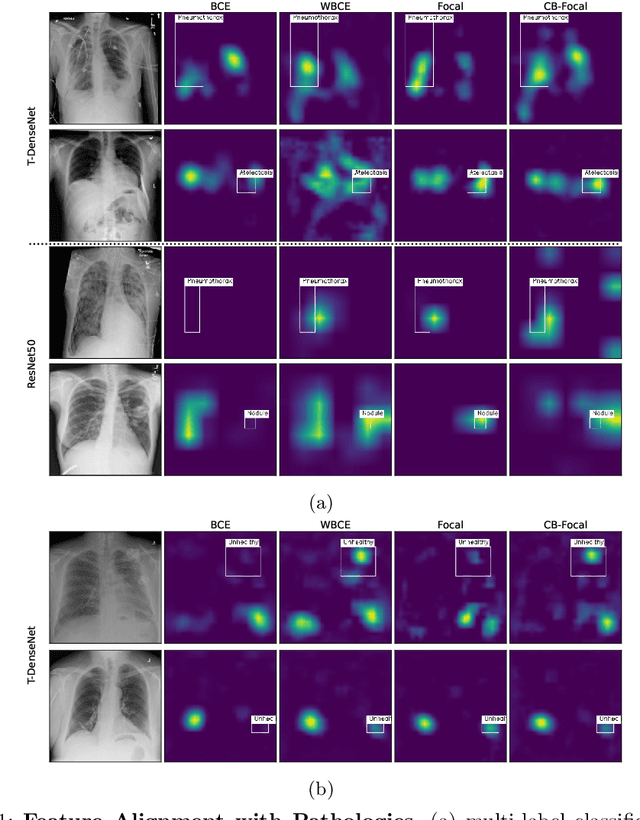
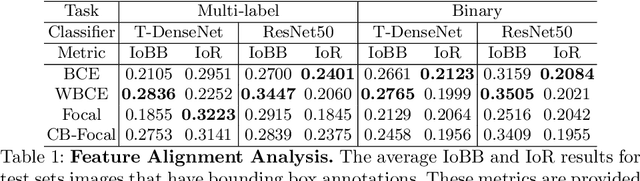
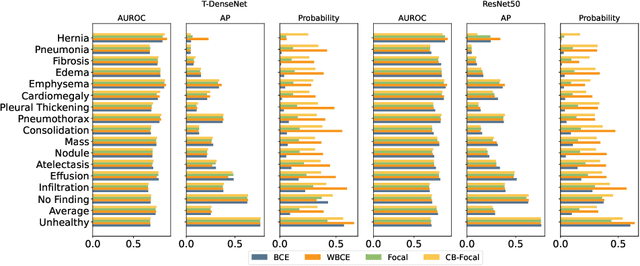
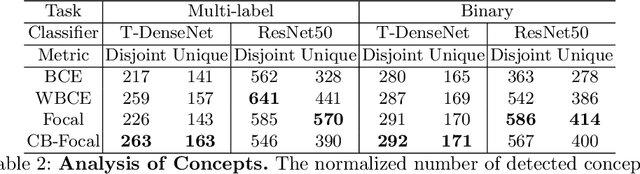
Abstract:One challenging property lurking in medical datasets is the imbalanced data distribution, where the frequency of the samples between the different classes is not balanced. Training a model on an imbalanced dataset can introduce unique challenges to the learning problem where a model is biased towards the highly frequent class. Many methods are proposed to tackle the distributional differences and the imbalanced problem. However, the impact of these approaches on the learned features is not well studied. In this paper, we look deeper into the internal units of neural networks to observe how handling data imbalance affects the learned features. We study several popular cost-sensitive approaches for handling data imbalance and analyze the feature maps of the convolutional neural networks from multiple perspectives: analyzing the alignment of salient features with pathologies and analyzing the pathology-related concepts encoded by the networks. Our study reveals differences and insights regarding the trained models that are not reflected by quantitative metrics such as AUROC and AP and show up only by looking at the models through a lens.
Positive-Unlabeled Learning with Uncertainty-aware Pseudo-label Selection
Jan 31, 2022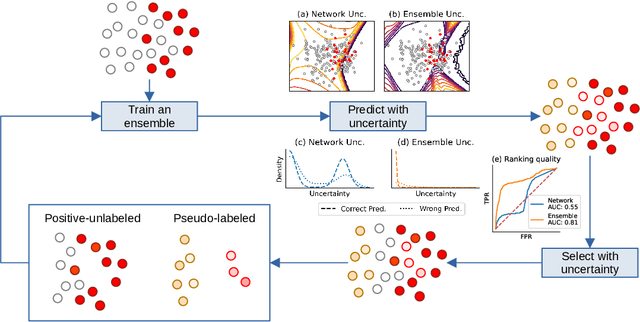
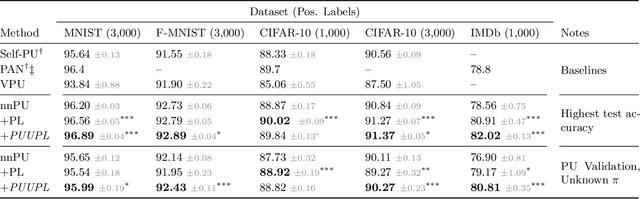
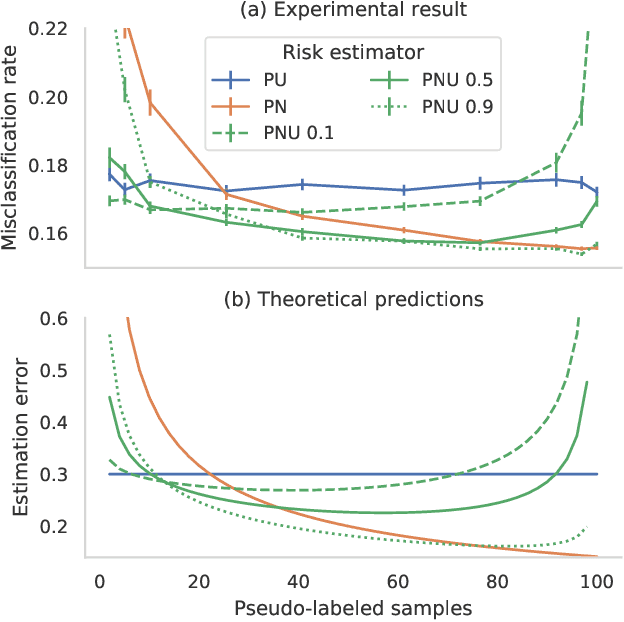
Abstract:Pseudo-labeling solutions for positive-unlabeled (PU) learning have the potential to result in higher performance compared to cost-sensitive learning but are vulnerable to incorrectly estimated pseudo-labels. In this paper, we provide a theoretical analysis of a risk estimator that combines risk on PU and pseudo-labeled data. Furthermore, we show analytically as well as experimentally that such an estimator results in lower excess risk compared to using PU data alone, provided that enough samples are pseudo-labeled with acceptable error rates. We then propose PUUPL, a novel training procedure for PU learning that leverages the epistemic uncertainty of an ensemble of deep neural networks to minimize errors in pseudo-label selection. We conclude with extensive experiments showing the effectiveness of our proposed algorithm over different datasets, modalities, and learning tasks. These show that PUUPL enables a reduction of up to 20% in test error rates even when prior and negative samples are not provided for validation, setting a new state-of-the-art for PU learning.
Deep Variational Clustering Framework for Self-labeling of Large-scale Medical Images
Sep 22, 2021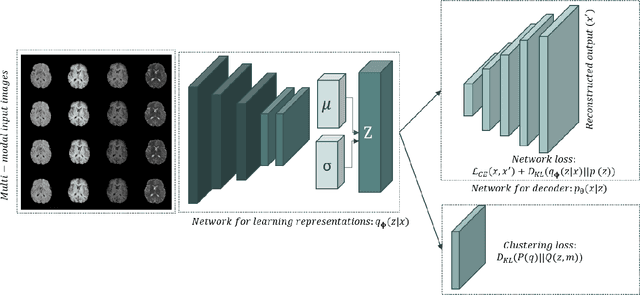
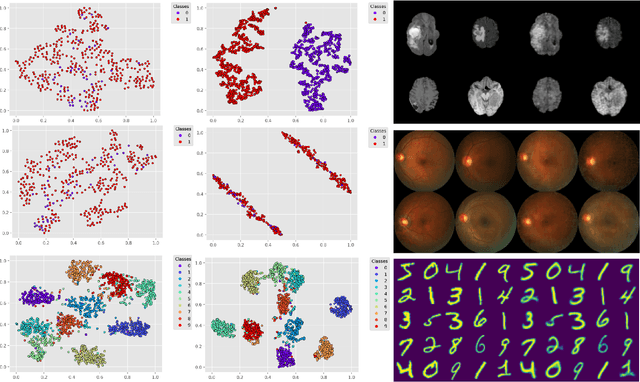
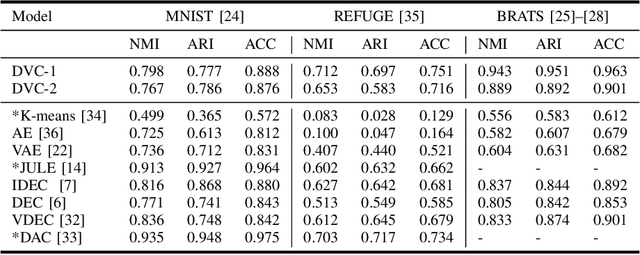
Abstract:We propose a Deep Variational Clustering (DVC) framework for unsupervised representation learning and clustering of large-scale medical images. DVC simultaneously learns the multivariate Gaussian posterior through the probabilistic convolutional encoder and the likelihood distribution with the probabilistic convolutional decoder; and optimizes cluster labels assignment. Here, the learned multivariate Gaussian posterior captures the latent distribution of a large set of unlabeled images. Then, we perform unsupervised clustering on top of the variational latent space using a clustering loss. In this approach, the probabilistic decoder helps to prevent the distortion of data points in the latent space and to preserve the local structure of data generating distribution. The training process can be considered as a self-training process to refine the latent space and simultaneously optimizing cluster assignments iteratively. We evaluated our proposed framework on three public datasets that represented different medical imaging modalities. Our experimental results show that our proposed framework generalizes better across different datasets. It achieves compelling results on several medical imaging benchmarks. Thus, our approach offers potential advantages over conventional deep unsupervised learning in real-world applications. The source code of the method and all the experiments are available publicly at: https://github.com/csfarzin/DVC
Deep Bregman Divergence for Contrastive Learning of Visual Representations
Sep 15, 2021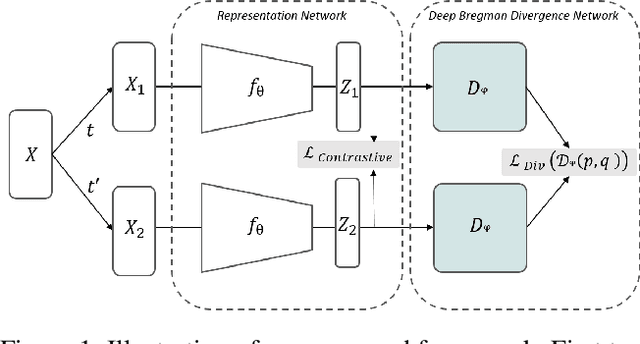
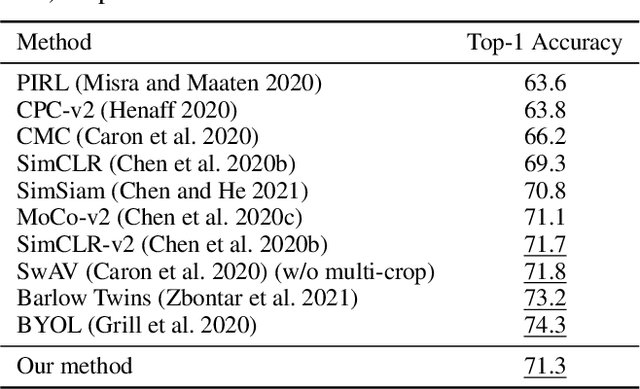
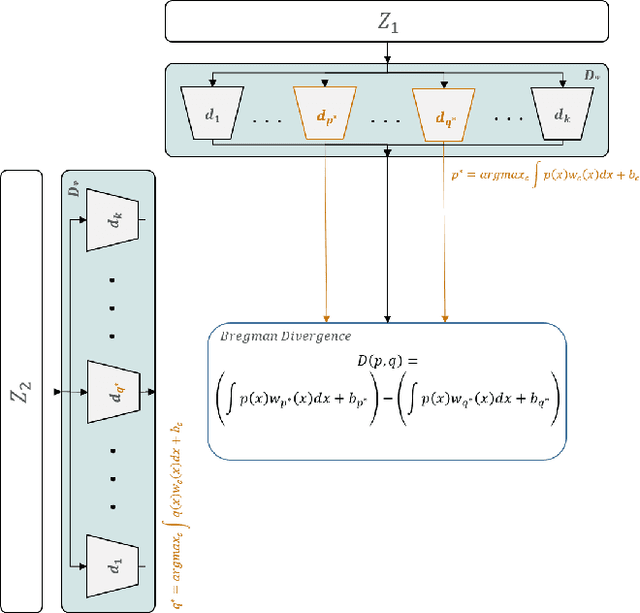
Abstract:Deep Bregman divergence measures divergence of data points using neural networks which is beyond Euclidean distance and capable of capturing divergence over distributions. In this paper, we propose deep Bregman divergences for contrastive learning of visual representation and we aim to enhance contrastive loss used in self-supervised learning by training additional networks based on functional Bregman divergence. In contrast to the conventional contrastive learning methods which are solely based on divergences between single points, our framework can capture the divergence between distributions which improves the quality of learned representation. By combining conventional contrastive loss with the proposed divergence loss, our method outperforms baseline and most of previous methods for self-supervised and semi-supervised learning on multiple classifications and object detection tasks and datasets. The source code of the method and of all the experiments are available at supplementary.
Learning Statistical Representation with Joint Deep Embedded Clustering
Sep 11, 2021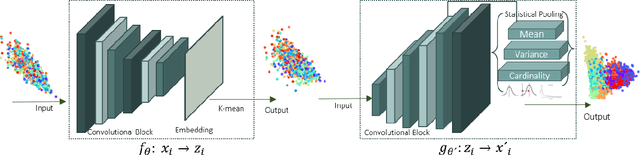
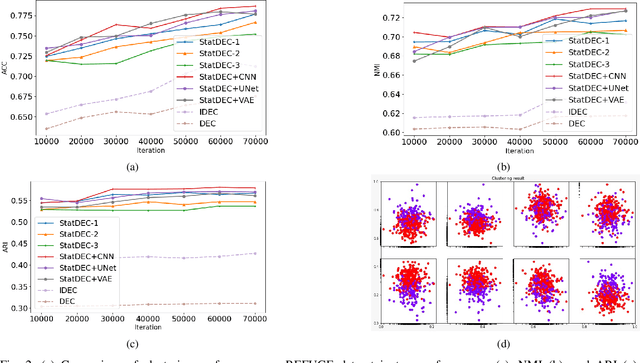
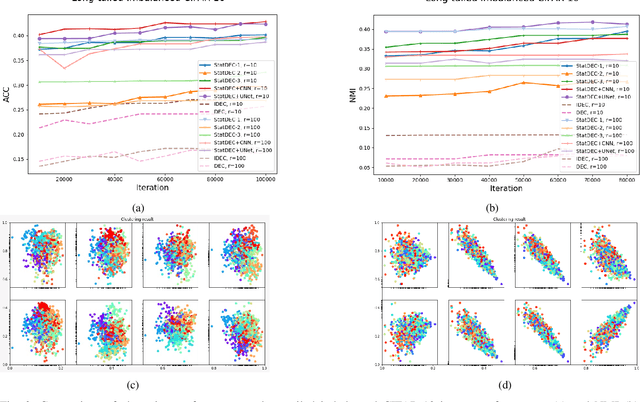
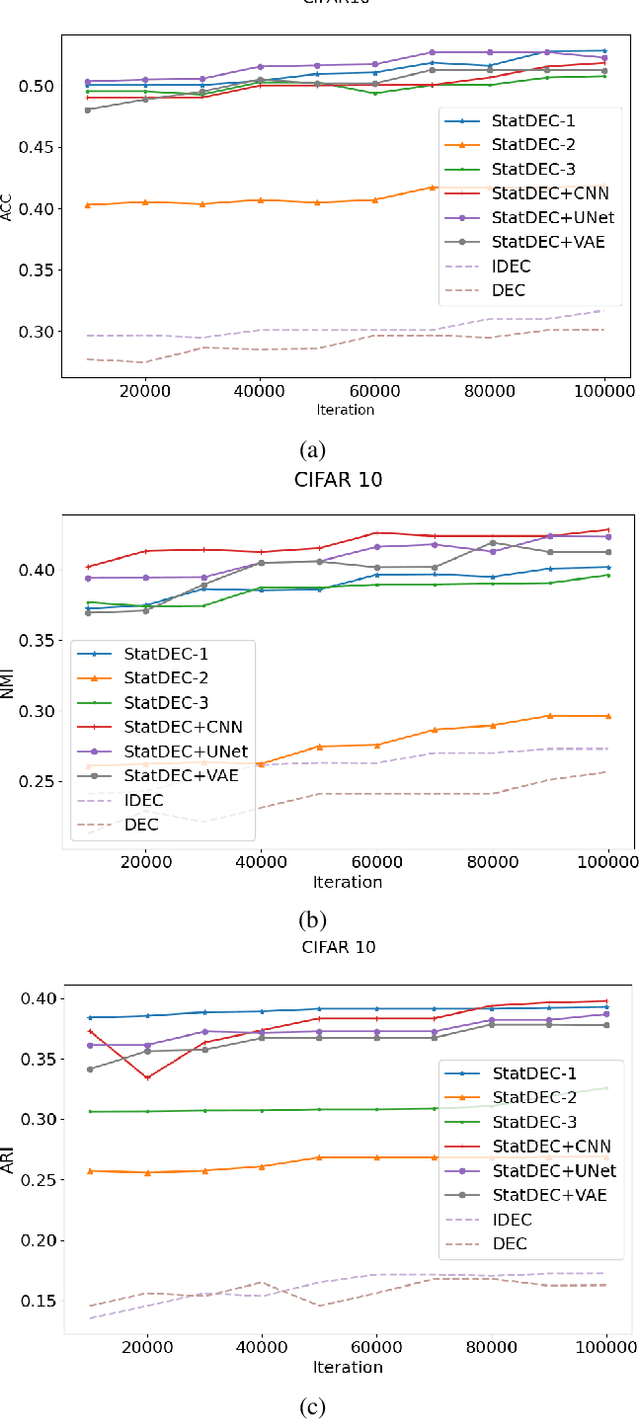
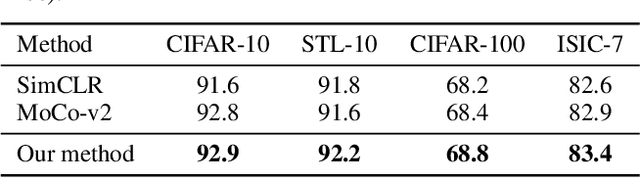
Abstract:One of the most promising approaches for unsupervised learning is combining deep representation learning and deep clustering. Some recent works propose to simultaneously learn representation using deep neural networks and perform clustering by defining a clustering loss on top of embedded features. However, these approaches are sensitive to imbalanced data and out-of-distribution samples. Hence, these methods optimize clustering by pushing data close to randomly initialized cluster centers. This is problematic when the number of instances varies largely in different classes or a cluster with few samples has less chance to be assigned a good centroid. To overcome these limitations, we introduce StatDEC, a new unsupervised framework for joint statistical representation learning and clustering. StatDEC simultaneously trains two deep learning models, a deep statistics network that captures the data distribution, and a deep clustering network that learns embedded features and performs clustering by explicitly defining a clustering loss. Specifically, the clustering network and representation network both take advantage of our proposed statistics pooling layer that represents mean, variance, and cardinality to handle the out-of-distribution samples as well as a class imbalance. Our experiments show that using these representations, one can considerably improve results on imbalanced image clustering across a variety of image datasets. Moreover, the learned representations generalize well when transferred to the out-of-distribution dataset.
Multi-Task Generative Adversarial Network for Handling Imbalanced Clinical Data
Nov 22, 2018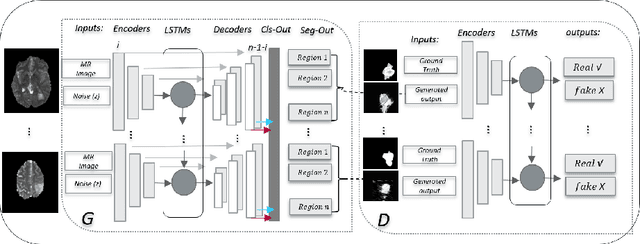
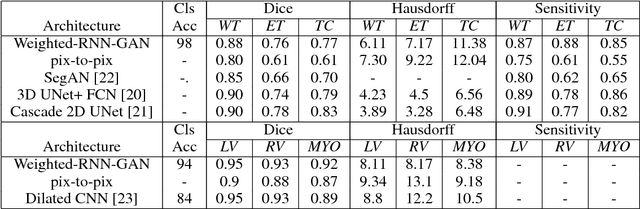
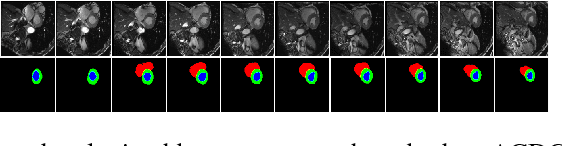
Abstract:We propose a new generative adversarial architecture to mitigate imbalance data problem for the task of medical image semantic segmentation where the majority of pixels belong to a healthy region and few belong to lesion or non-health region. A model trained with imbalanced data tends to bias towards healthy data which is not desired in clinical applications. We design a new conditional GAN with two components: a generative model and a discriminative model to mitigate imbalanced data problem through selective weighted loss. While the generator is trained on sequential magnetic resonance images (MRI) to learn semantic segmentation and disease classification, the discriminator classifies whether a generated output is real or fake. The proposed architecture achieved state-of-the-art results on ACDC-2017 for cardiac segmentation and diseases classification. We have achieved competitive results on BraTS-2017 for brain tumor segmentation and brain diseases classification.
Conditional Generative Refinement Adversarial Networks for Unbalanced Medical Image Semantic Segmentation
Oct 09, 2018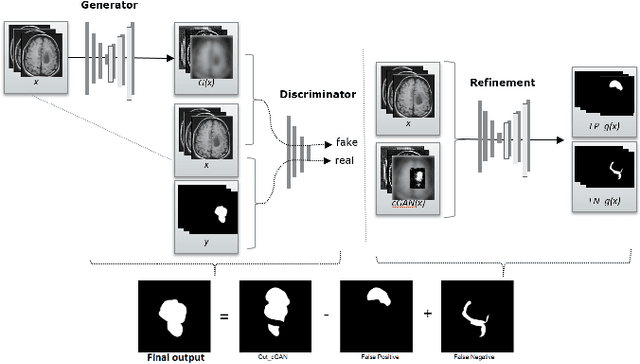
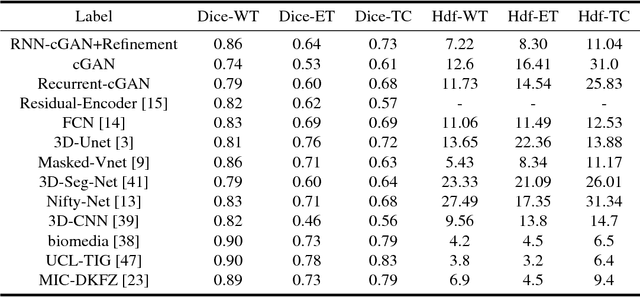
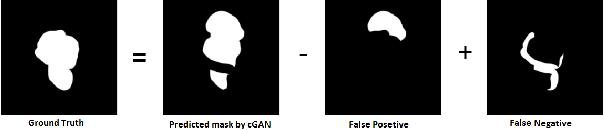
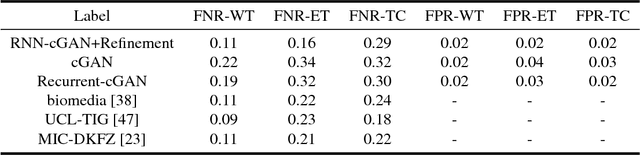
Abstract:We propose a new generative adversarial architecture to mitigate imbalance data problem in medical image semantic segmentation where the majority of pixels belongs to a healthy region and few belong to lesion or non-health region. A model trained with imbalanced data tends to bias toward healthy data which is not desired in clinical applications and predicted outputs by these networks have high precision and low sensitivity. We propose a new conditional generative refinement network with three components: a generative, a discriminative, and a refinement network to mitigate unbalanced data problem through ensemble learning. The generative network learns to a segment at the pixel level by getting feedback from the discriminative network according to the true positive and true negative maps. On the other hand, the refinement network learns to predict the false positive and the false negative masks produced by the generative network that has significant value, especially in medical application. The final semantic segmentation masks are then composed by the output of the three networks. The proposed architecture shows state-of-the-art results on LiTS-2017 for liver lesion segmentation, and two microscopic cell segmentation datasets MDA231, PhC-HeLa. We have achieved competitive results on BraTS-2017 for brain tumour segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge