Why is the winner the best?
Mar 30, 2023Matthias Eisenmann, Annika Reinke, Vivienn Weru, Minu Dietlinde Tizabi, Fabian Isensee, Tim J. Adler, Sharib Ali, Vincent Andrearczyk, Marc Aubreville, Ujjwal Baid, Spyridon Bakas, Niranjan Balu, Sophia Bano, Jorge Bernal, Sebastian Bodenstedt, Alessandro Casella, Veronika Cheplygina, Marie Daum, Marleen de Bruijne, Adrien Depeursinge, Reuben Dorent, Jan Egger, David G. Ellis, Sandy Engelhardt, Melanie Ganz, Noha Ghatwary, Gabriel Girard, Patrick Godau, Anubha Gupta, Lasse Hansen, Kanako Harada, Mattias Heinrich, Nicholas Heller, Alessa Hering, Arnaud Huaulmé, Pierre Jannin, Ali Emre Kavur, Oldřich Kodym, Michal Kozubek, Jianning Li, Hongwei Li, Jun Ma, Carlos Martín-Isla, Bjoern Menze, Alison Noble, Valentin Oreiller, Nicolas Padoy, Sarthak Pati, Kelly Payette, Tim Rädsch, Jonathan Rafael-Patiño, Vivek Singh Bawa, Stefanie Speidel, Carole H. Sudre, Kimberlin van Wijnen, Martin Wagner, Donglai Wei, Amine Yamlahi, Moi Hoon Yap, Chun Yuan, Maximilian Zenk, Aneeq Zia, David Zimmerer, Dogu Baran Aydogan, Binod Bhattarai, Louise Bloch, Raphael Brüngel, Jihoon Cho, Chanyeol Choi, Qi Dou, Ivan Ezhov, Christoph M. Friedrich, Clifton Fuller, Rebati Raman Gaire, Adrian Galdran, Álvaro García Faura, Maria Grammatikopoulou, SeulGi Hong, Mostafa Jahanifar, Ikbeom Jang, Abdolrahim Kadkhodamohammadi, Inha Kang, Florian Kofler, Satoshi Kondo, Hugo Kuijf, Mingxing Li, Minh Huan Luu, Tomaž Martinčič, Pedro Morais, Mohamed A. Naser, Bruno Oliveira, David Owen, Subeen Pang, Jinah Park, Sung-Hong Park, Szymon Płotka, Elodie Puybareau, Nasir Rajpoot, Kanghyun Ryu, Numan Saeed, Adam Shephard, Pengcheng Shi, Dejan Štepec, Ronast Subedi, Guillaume Tochon, Helena R. Torres, Helene Urien, João L. Vilaça, Kareem Abdul Wahid, Haojie Wang, Jiacheng Wang, Liansheng Wang, Xiyue Wang, Benedikt Wiestler, Marek Wodzinski, Fangfang Xia, Juanying Xie, Zhiwei Xiong, Sen Yang, Yanwu Yang, Zixuan Zhao, Klaus Maier-Hein, Paul F. Jäger, Annette Kopp-Schneider, Lena Maier-Hein
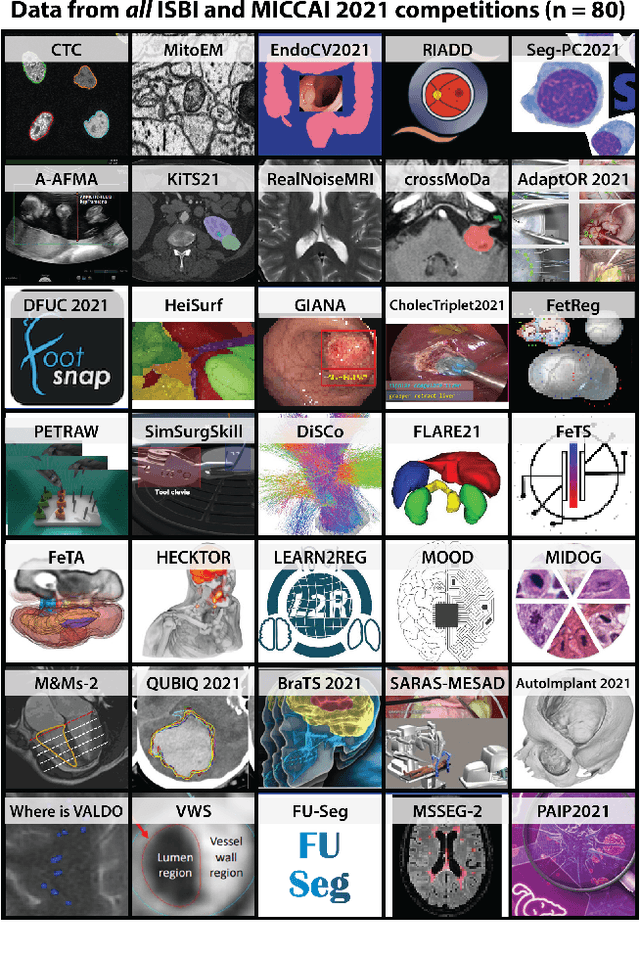
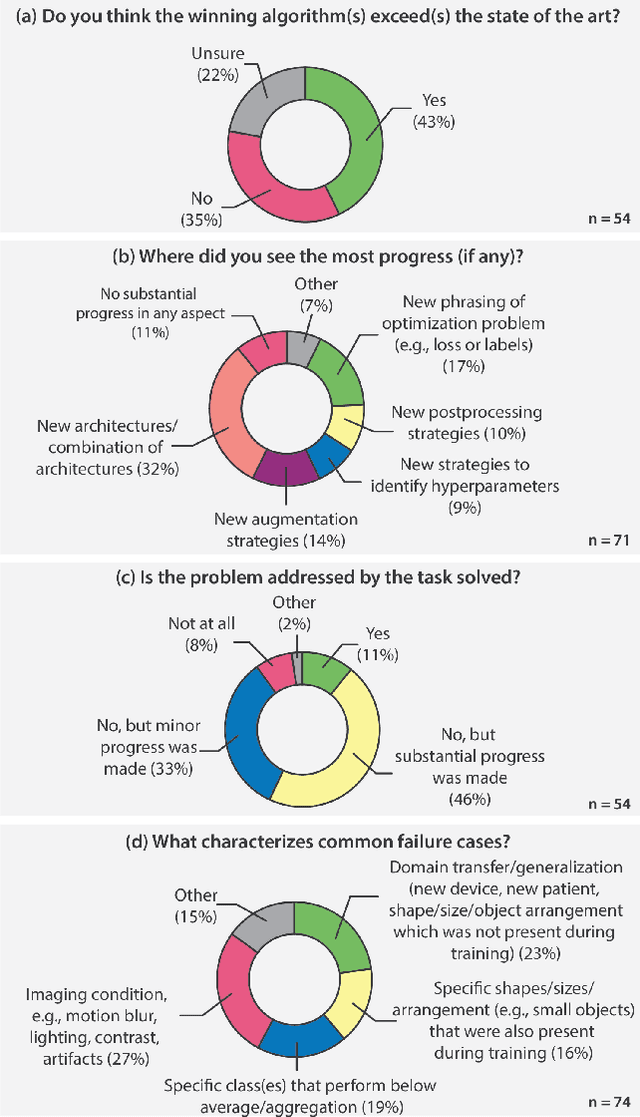
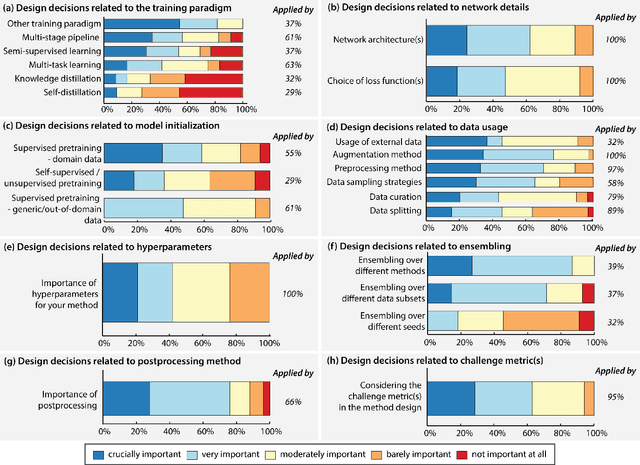
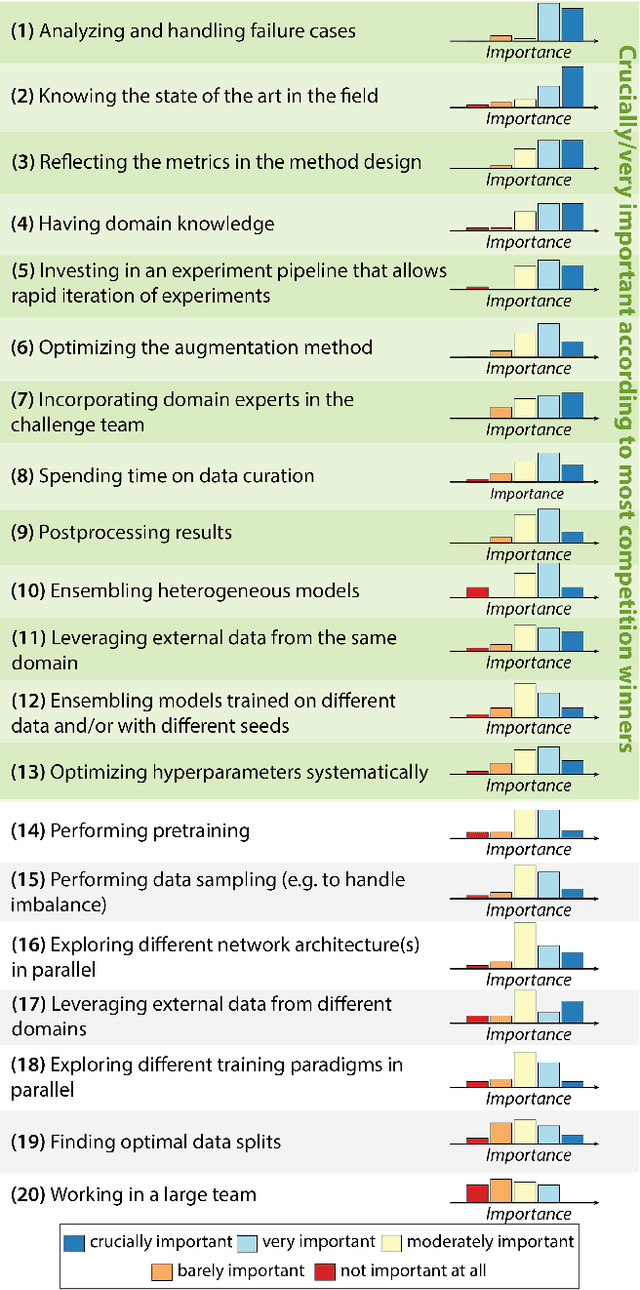
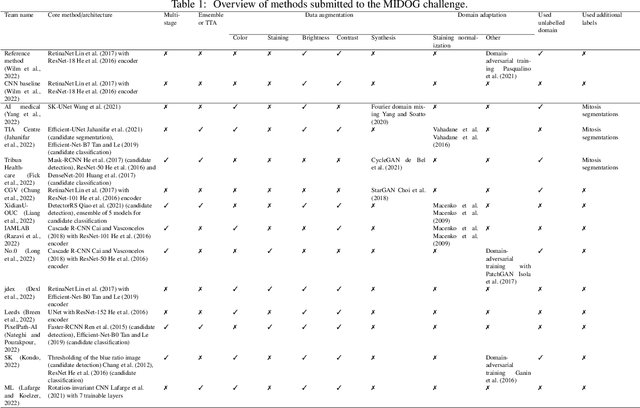
International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Multi-Scanner Canine Cutaneous Squamous Cell Carcinoma Histopathology Dataset
Jan 11, 2023Frauke Wilm, Marco Fragoso, Christof A. Bertram, Nikolas Stathonikos, Mathias Öttl, Jingna Qiu, Robert Klopfleisch, Andreas Maier, Katharina Breininger, Marc Aubreville
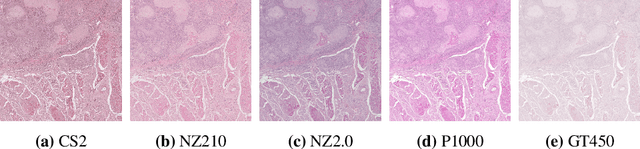
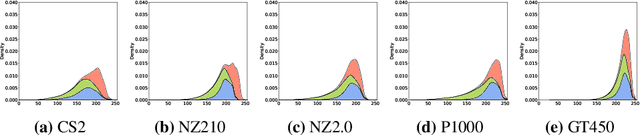
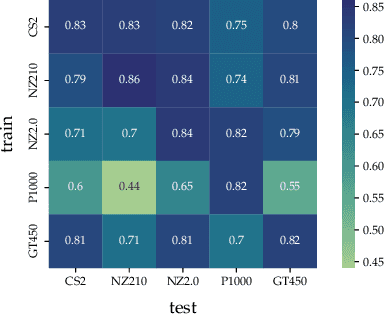
In histopathology, scanner-induced domain shifts are known to impede the performance of trained neural networks when tested on unseen data. Multi-domain pre-training or dedicated domain-generalization techniques can help to develop domain-agnostic algorithms. For this, multi-scanner datasets with a high variety of slide scanning systems are highly desirable. We present a publicly available multi-scanner dataset of canine cutaneous squamous cell carcinoma histopathology images, composed of 44 samples digitized with five slide scanners. This dataset provides local correspondences between images and thereby isolates the scanner-induced domain shift from other inherent, e.g. morphology-induced domain shifts. To highlight scanner differences, we present a detailed evaluation of color distributions, sharpness, and contrast of the individual scanner subsets. Additionally, to quantify the inherent scanner-induced domain shift, we train a tumor segmentation network on each scanner subset and evaluate the performance both in- and cross-domain. We achieve a class-averaged in-domain intersection over union coefficient of up to 0.86 and observe a cross-domain performance decrease of up to 0.38, which confirms the inherent domain shift of the presented dataset and its negative impact on the performance of deep neural networks.
Attention-based Multiple Instance Learning for Survival Prediction on Lung Cancer Tissue Microarrays
Dec 15, 2022Jonas Ammeling, Lars-Henning Schmidt, Jonathan Ganz, Tanja Niedermair, Christoph Brochhausen-Delius, Christian Schulz, Katharina Breininger, Marc Aubreville
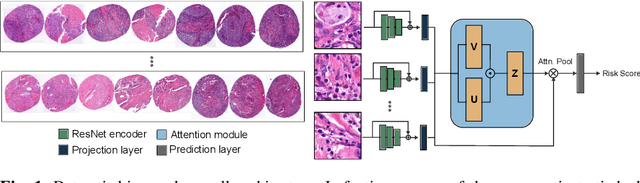
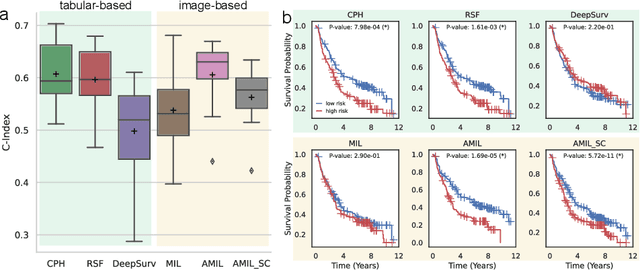
Attention-based multiple instance learning (AMIL) algorithms have proven to be successful in utilizing gigapixel whole-slide images (WSIs) for a variety of different computational pathology tasks such as outcome prediction and cancer subtyping problems. We extended an AMIL approach to the task of survival prediction by utilizing the classical Cox partial likelihood as a loss function, converting the AMIL model into a nonlinear proportional hazards model. We applied the model to tissue microarray (TMA) slides of 330 lung cancer patients. The results show that AMIL approaches can handle very small amounts of tissue from a TMA and reach similar C-index performance compared to established survival prediction methods trained with highly discriminative clinical factors such as age, cancer grade, and cancer stage
Deep Learning-Based Automatic Assessment of AgNOR-scores in Histopathology Images
Dec 15, 2022Jonathan Ganz, Karoline Lipnik, Jonas Ammeling, Barbara Richter, Chloé Puget, Eda Parlak, Laura Diehl, Robert Klopfleisch, Taryn A. Donovan, Matti Kiupel, Christof A. Bertram, Katharina Breininger, Marc Aubreville
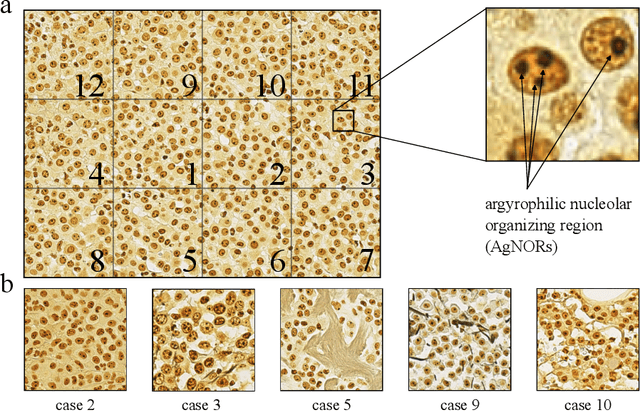
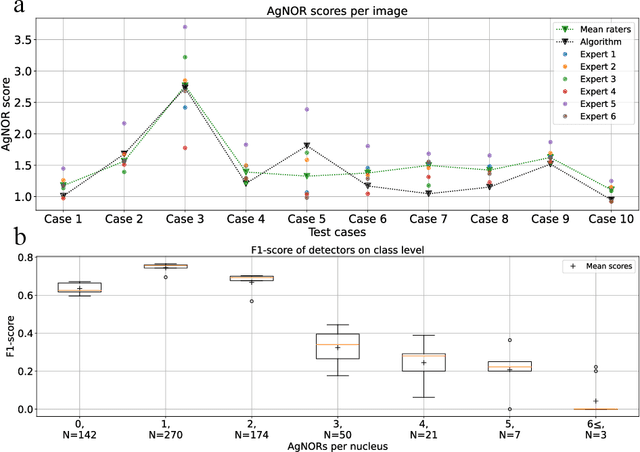
Nucleolar organizer regions (NORs) are parts of the DNA that are involved in RNA transcription. Due to the silver affinity of associated proteins, argyrophilic NORs (AgNORs) can be visualized using silver-based staining. The average number of AgNORs per nucleus has been shown to be a prognostic factor for predicting the outcome of many tumors. Since manual detection of AgNORs is laborious, automation is of high interest. We present a deep learning-based pipeline for automatically determining the AgNOR-score from histopathological sections. An additional annotation experiment was conducted with six pathologists to provide an independent performance evaluation of our approach. Across all raters and images, we found a mean squared error of 0.054 between the AgNOR- scores of the experts and those of the model, indicating that our approach offers performance comparable to humans.
Deep learning-based Subtyping of Atypical and Normal Mitoses using a Hierarchical Anchor-Free Object Detector
Dec 12, 2022Marc Aubreville, Jonathan Ganz, Jonas Ammeling, Taryn A. Donovan, Rutger H. J. Fick, Katharina Breininger, Christof A. Bertram
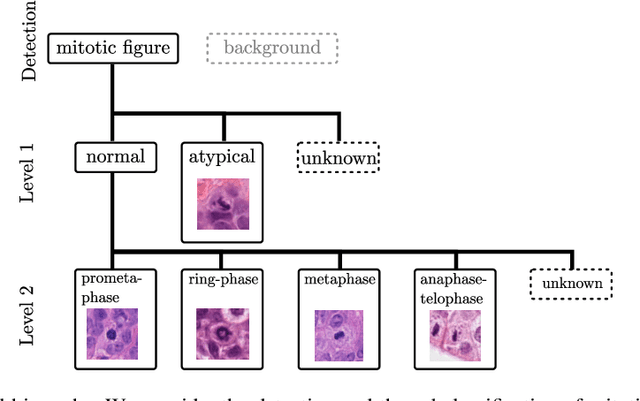
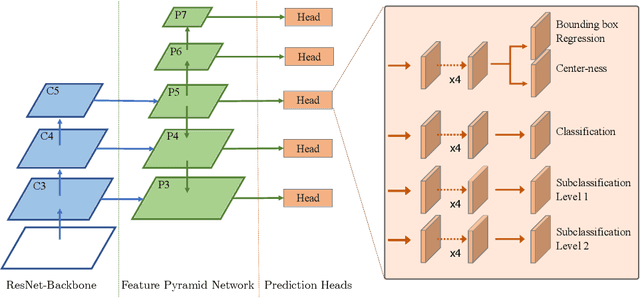
Mitotic activity is key for the assessment of malignancy in many tumors. Moreover, it has been demonstrated that the proportion of abnormal mitosis to normal mitosis is of prognostic significance. Atypical mitotic figures (MF) can be identified morphologically as having segregation abnormalities of the chromatids. In this work, we perform, for the first time, automatic subtyping of mitotic figures into normal and atypical categories according to characteristic morphological appearances of the different phases of mitosis. Using the publicly available MIDOG21 and TUPAC16 breast cancer mitosis datasets, two experts blindly subtyped mitotic figures into five morphological categories. Further, we set up a state-of-the-art object detection pipeline extending the anchor-free FCOS approach with a gated hierarchical subclassification branch. Our labeling experiment indicated that subtyping of mitotic figures is a challenging task and prone to inter-rater disagreement, which we found in 24.89% of MF. Using the more diverse MIDOG21 dataset for training and TUPAC16 for testing, we reached a mean overall average precision score of 0.552, a ROC AUC score of 0.833 for atypical/normal MF and a mean class-averaged ROC-AUC score of 0.977 for discriminating the different phases of cells undergoing mitosis.
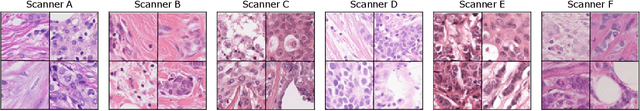
Mind the Gap: Scanner-induced domain shifts pose challenges for representation learning in histopathology
Nov 29, 2022Frauke Wilm, Marco Fragoso, Christof A. Bertram, Nikolas Stathonikos, Mathias Öttl, Jingna Qiu, Robert Klopfleisch, Andreas Maier, Marc Aubreville, Katharina Breininger
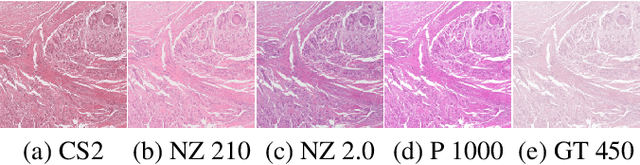
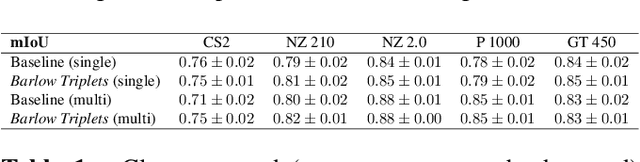
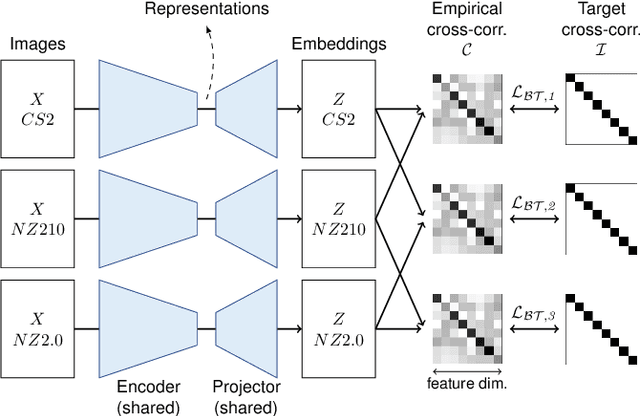
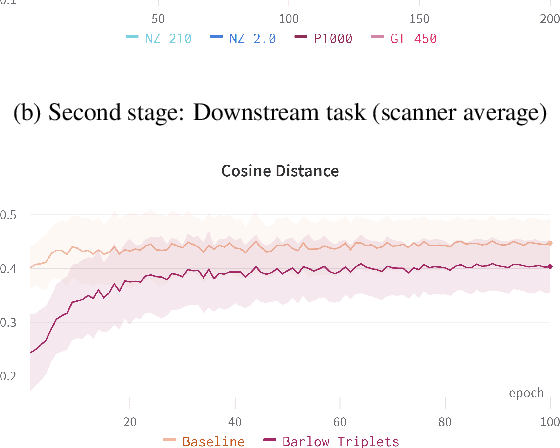
Computer-aided systems in histopathology are often challenged by various sources of domain shift that impact the performance of these algorithms considerably. We investigated the potential of using self-supervised pre-training to overcome scanner-induced domain shifts for the downstream task of tumor segmentation. For this, we present the Barlow Triplets to learn scanner-invariant representations from a multi-scanner dataset with local image correspondences. We show that self-supervised pre-training successfully aligned different scanner representations, which, interestingly only results in a limited benefit for our downstream task. We thereby provide insights into the influence of scanner characteristics for downstream applications and contribute to a better understanding of why established self-supervised methods have not yet shown the same success on histopathology data as they have for natural images.
Fast Text-Conditional Discrete Denoising on Vector-Quantized Latent Spaces
Nov 14, 2022Dominic Rampas, Pablo Pernias, Elea Zhong, Marc Aubreville
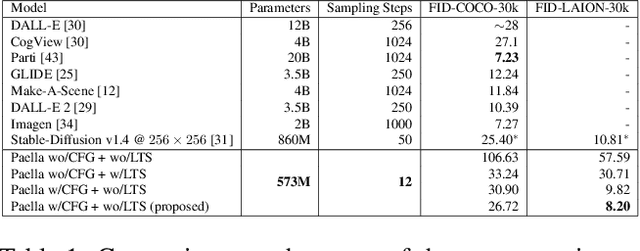
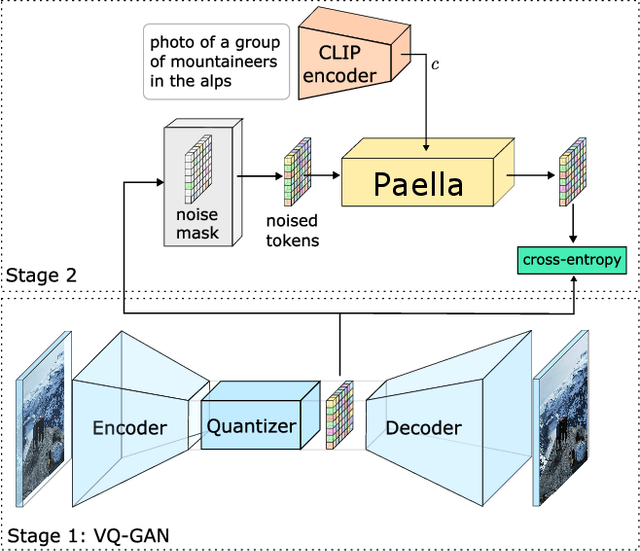
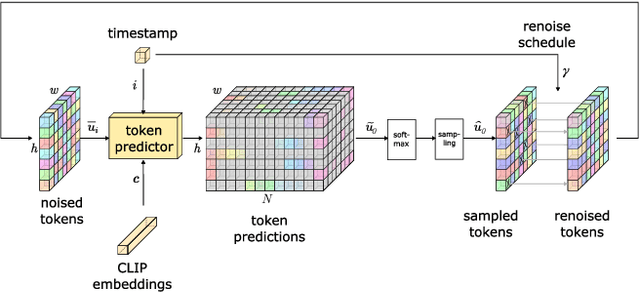
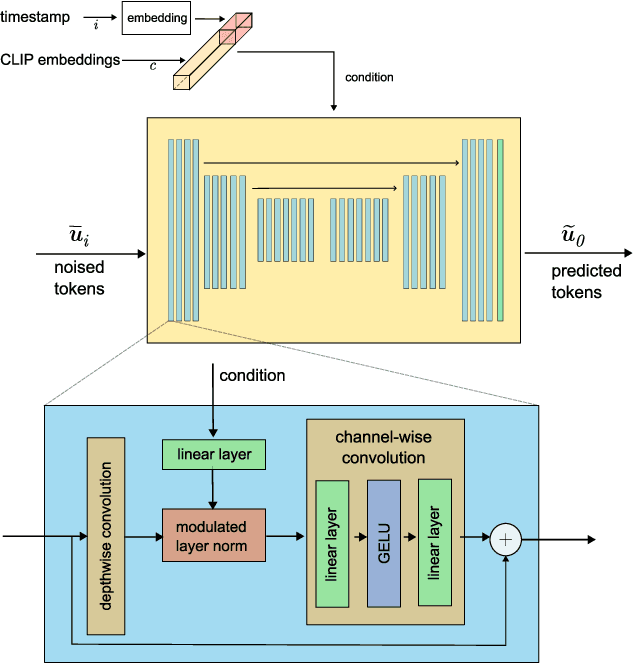
Conditional text-to-image generation has seen countless recent improvements in terms of quality, diversity and fidelity. Nevertheless, most state-of-the-art models require numerous inference steps to produce faithful generations, resulting in performance bottlenecks for end-user applications. In this paper we introduce Paella, a novel text-to-image model requiring less than 10 steps to sample high-fidelity images, using a speed-optimized architecture allowing to sample a single image in less than 500 ms, while having 573M parameters. The model operates on a compressed & quantized latent space, it is conditioned on CLIP embeddings and uses an improved sampling function over previous works. Aside from text-conditional image generation, our model is able to do latent space interpolation and image manipulations such as inpainting, outpainting, and structural editing. We release all of our code and pretrained models at https://github.com/dome272/Paella
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022Marc Aubreville, Nikolas Stathonikos, Christof A. Bertram, Robert Klopleisch, Natalie ter Hoeve, Francesco Ciompi, Frauke Wilm, Christian Marzahl, Taryn A. Donovan, Andreas Maier, Jack Breen, Nishant Ravikumar, Youjin Chung, Jinah Park, Ramin Nateghi, Fattaneh Pourakpour, Rutger H. J. Fick, Saima Ben Hadj, Mostafa Jahanifar, Nasir Rajpoot, Jakob Dexl, Thomas Wittenberg, Satoshi Kondo, Maxime W. Lafarge, Viktor H. Koelzer, Jingtang Liang, Yubo Wang, Xi Long, Jingxin Liu, Salar Razavi, April Khademi, Sen Yang, Xiyue Wang, Mitko Veta, Katharina Breininger
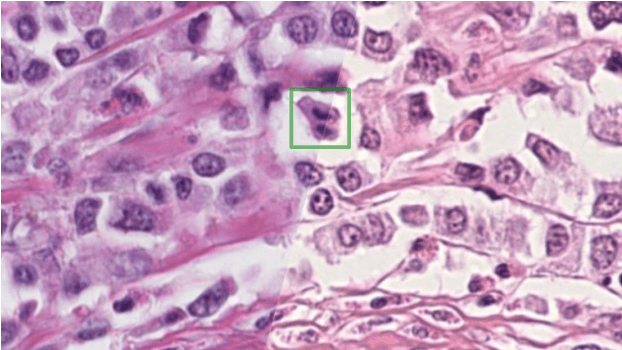
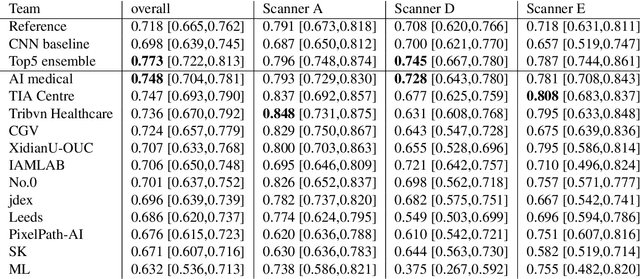
The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
Pan-Tumor CAnine cuTaneous Cancer Histology (CATCH) Dataset
Jan 27, 2022Frauke Wilm, Marco Fragoso, Christian Marzahl, Jingna Qiu, Christof A. Bertram, Robert Klopfleisch, Andreas Maier, Katharina Breininger, Marc Aubreville
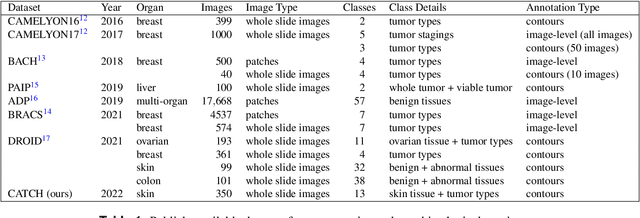
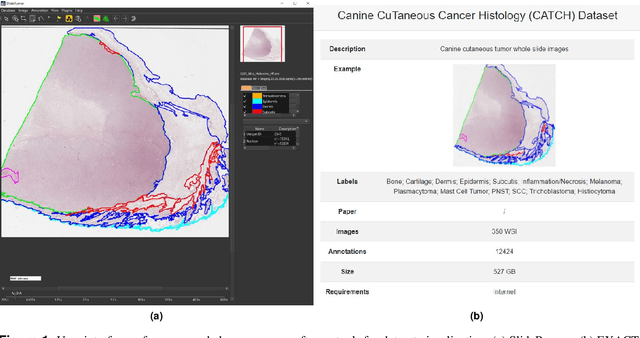
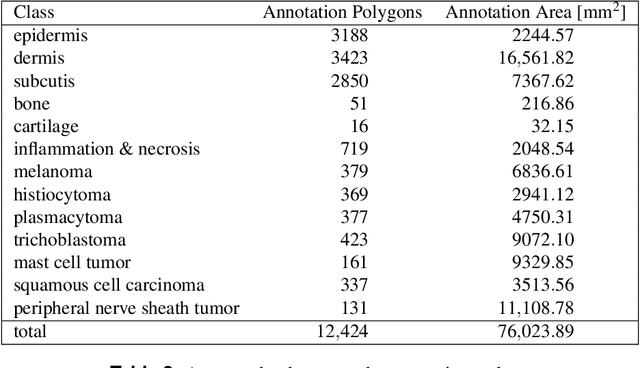
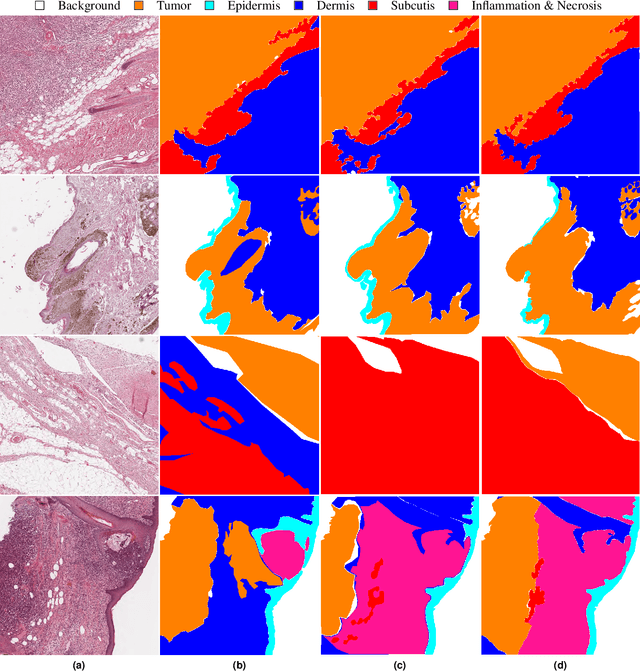
Due to morphological similarities, the differentiation of histologic sections of cutaneous tumors into individual subtypes can be challenging. Recently, deep learning-based approaches have proven their potential for supporting pathologists in this regard. However, many of these supervised algorithms require a large amount of annotated data for robust development. We present a publicly available dataset consisting of 350 whole slide images of seven different canine cutaneous tumors complemented by 12,424 polygon annotations for 13 histologic classes including seven cutaneous tumor subtypes. Regarding sample size and annotation extent, this exceeds most publicly available datasets which are oftentimes limited to the tumor area or merely provide patch-level annotations. We validated our model for tissue segmentation, achieving a class-averaged Jaccard coefficient of 0.7047, and 0.9044 for tumor in particular. For tumor subtype classification, we achieve a slide-level accuracy of 0.9857. Since canine cutaneous tumors possess various histologic homologies to human tumors, we believe that the added value of this dataset is not limited to veterinary pathology but extends to more general fields of application.
Domain Adversarial RetinaNet as a Reference Algorithm for the MItosis DOmain Generalization (MIDOG) Challenge
Aug 25, 2021Frauke Wilm, Katharina Breininger, Marc Aubreville
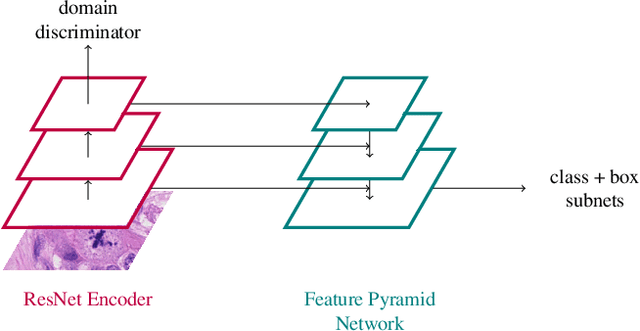
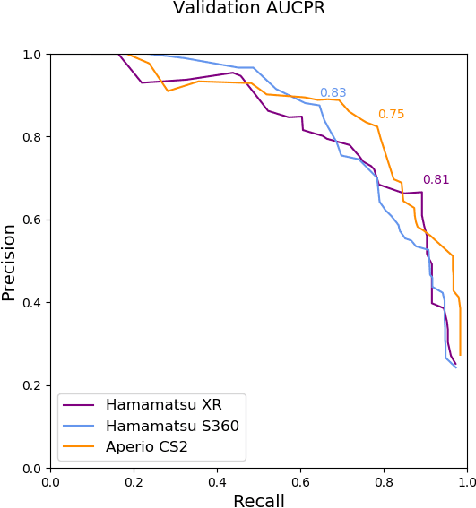
Assessing the Mitotic Count has a known high degree of intra- and inter-rater variability. Computer-aided systems have proven to decrease this variability and reduce labelling time. These systems, however, are generally highly dependent on their training domain and show poor applicability to unseen domains. In histopathology, these domain shifts can result from various sources, including different slide scanning systems used to digitize histologic samples. The MItosis DOmain Generalization challenge focuses on this specific domain shift for the task of mitotic figure detection. This work presents a mitotic figure detection algorithm developed as a baseline for the challenge, based on domain adversarial training. On the preliminary test set, the algorithm scores an F$_1$ score of 0.7514.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge