Lisa Adams
Agentic large language models improve retrieval-based radiology question answering
Aug 01, 2025



Abstract:Clinical decision-making in radiology increasingly benefits from artificial intelligence (AI), particularly through large language models (LLMs). However, traditional retrieval-augmented generation (RAG) systems for radiology question answering (QA) typically rely on single-step retrieval, limiting their ability to handle complex clinical reasoning tasks. Here we propose an agentic RAG framework enabling LLMs to autonomously decompose radiology questions, iteratively retrieve targeted clinical evidence from Radiopaedia, and dynamically synthesize evidence-based responses. We evaluated 24 LLMs spanning diverse architectures, parameter scales (0.5B to >670B), and training paradigms (general-purpose, reasoning-optimized, clinically fine-tuned), using 104 expert-curated radiology questions from previously established RSNA-RadioQA and ExtendedQA datasets. Agentic retrieval significantly improved mean diagnostic accuracy over zero-shot prompting (73% vs. 64%; P<0.001) and conventional online RAG (73% vs. 68%; P<0.001). The greatest gains occurred in mid-sized models (e.g., Mistral Large improved from 72% to 81%) and small-scale models (e.g., Qwen 2.5-7B improved from 55% to 71%), while very large models (>200B parameters) demonstrated minimal changes (<2% improvement). Additionally, agentic retrieval reduced hallucinations (mean 9.4%) and retrieved clinically relevant context in 46% of cases, substantially aiding factual grounding. Even clinically fine-tuned models exhibited meaningful improvements (e.g., MedGemma-27B improved from 71% to 81%), indicating complementary roles of retrieval and fine-tuning. These results highlight the potential of agentic frameworks to enhance factuality and diagnostic accuracy in radiology QA, particularly among mid-sized LLMs, warranting future studies to validate their clinical utility.
LongHealth: A Question Answering Benchmark with Long Clinical Documents
Jan 25, 2024



Abstract:Background: Recent advancements in large language models (LLMs) offer potential benefits in healthcare, particularly in processing extensive patient records. However, existing benchmarks do not fully assess LLMs' capability in handling real-world, lengthy clinical data. Methods: We present the LongHealth benchmark, comprising 20 detailed fictional patient cases across various diseases, with each case containing 5,090 to 6,754 words. The benchmark challenges LLMs with 400 multiple-choice questions in three categories: information extraction, negation, and sorting, challenging LLMs to extract and interpret information from large clinical documents. Results: We evaluated nine open-source LLMs with a minimum of 16,000 tokens and also included OpenAI's proprietary and cost-efficient GPT-3.5 Turbo for comparison. The highest accuracy was observed for Mixtral-8x7B-Instruct-v0.1, particularly in tasks focused on information retrieval from single and multiple patient documents. However, all models struggled significantly in tasks requiring the identification of missing information, highlighting a critical area for improvement in clinical data interpretation. Conclusion: While LLMs show considerable potential for processing long clinical documents, their current accuracy levels are insufficient for reliable clinical use, especially in scenarios requiring the identification of missing information. The LongHealth benchmark provides a more realistic assessment of LLMs in a healthcare setting and highlights the need for further model refinement for safe and effective clinical application. We make the benchmark and evaluation code publicly available.
From Text to Image: Exploring GPT-4Vision's Potential in Advanced Radiological Analysis across Subspecialties
Nov 24, 2023

Abstract:The study evaluates and compares GPT-4 and GPT-4Vision for radiological tasks, suggesting GPT-4Vision may recognize radiological features from images, thereby enhancing its diagnostic potential over text-based descriptions.
Comparing Different Deep Learning Architectures for Classification of Chest Radiographs
Feb 20, 2020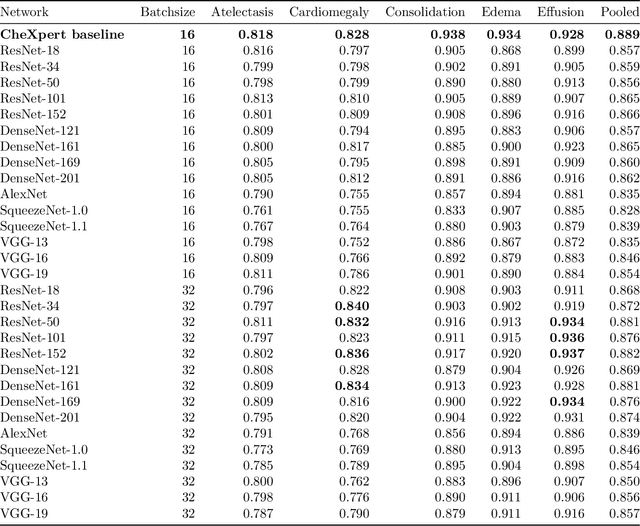
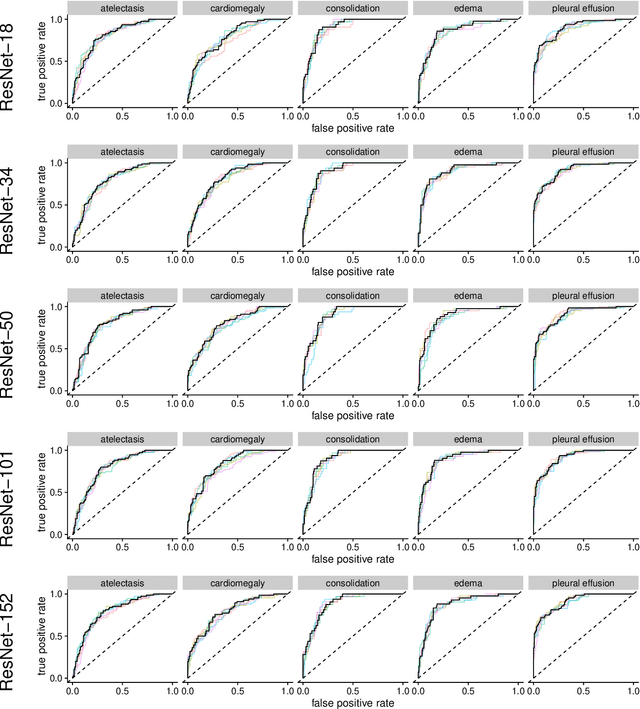
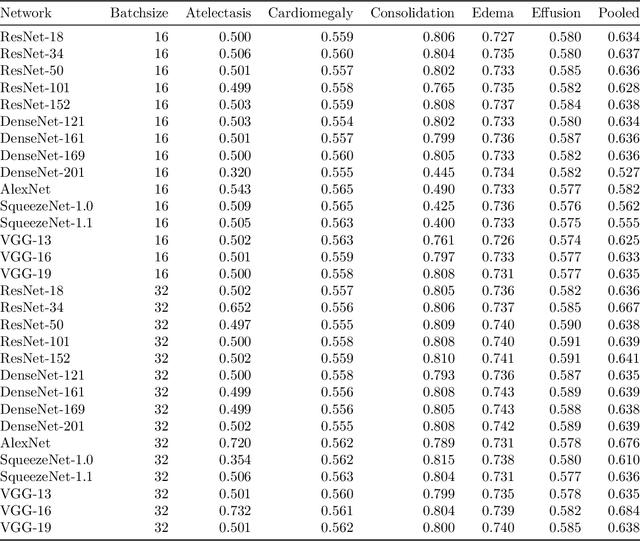
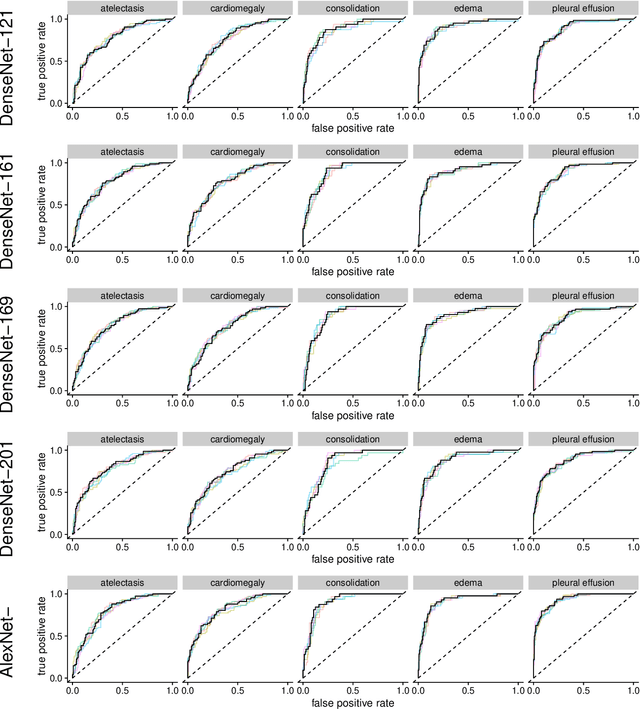
Abstract:Chest radiographs are among the most frequently acquired images in radiology and are often the subject of computer vision research. However, most of the models used to classify chest radiographs are derived from openly available deep neural networks, trained on large image-datasets. These datasets routinely differ from chest radiographs in that they are mostly color images and contain several possible image classes, while radiographs are greyscale images and often only contain fewer image classes. Therefore, very deep neural networks, which can represent more complex relationships in image-features, might not be required for the comparatively simpler task of classifying grayscale chest radiographs. We compared fifteen different architectures of artificial neural networks regarding training-time and performance on the openly available CheXpert dataset to identify the most suitable models for deep learning tasks on chest radiographs. We could show, that smaller networks such as ResNet-34, AlexNet or VGG-16 have the potential to classify chest radiographs as precisely as deeper neural networks such as DenseNet-201 or ResNet-151, while being less computationally demanding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge