Lily H. Peng
Predicting Prostate Cancer-Specific Mortality with A.I.-based Gleason Grading
Nov 25, 2020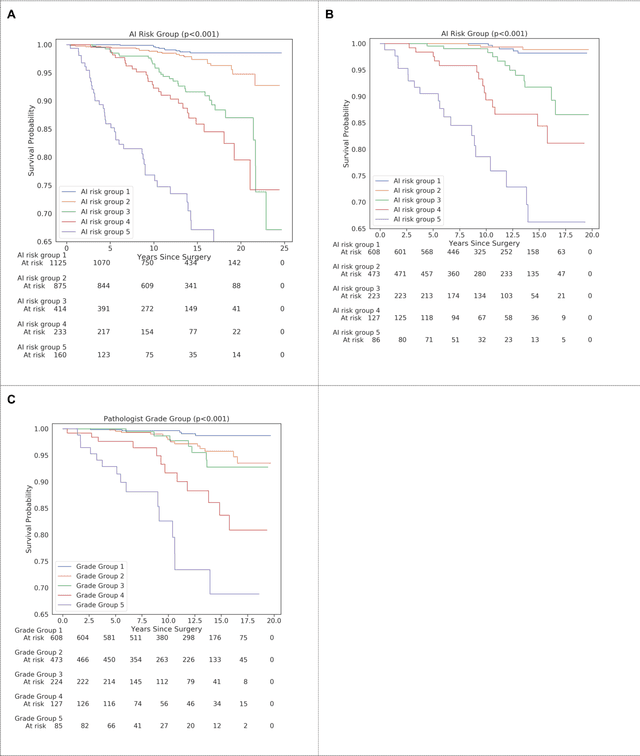
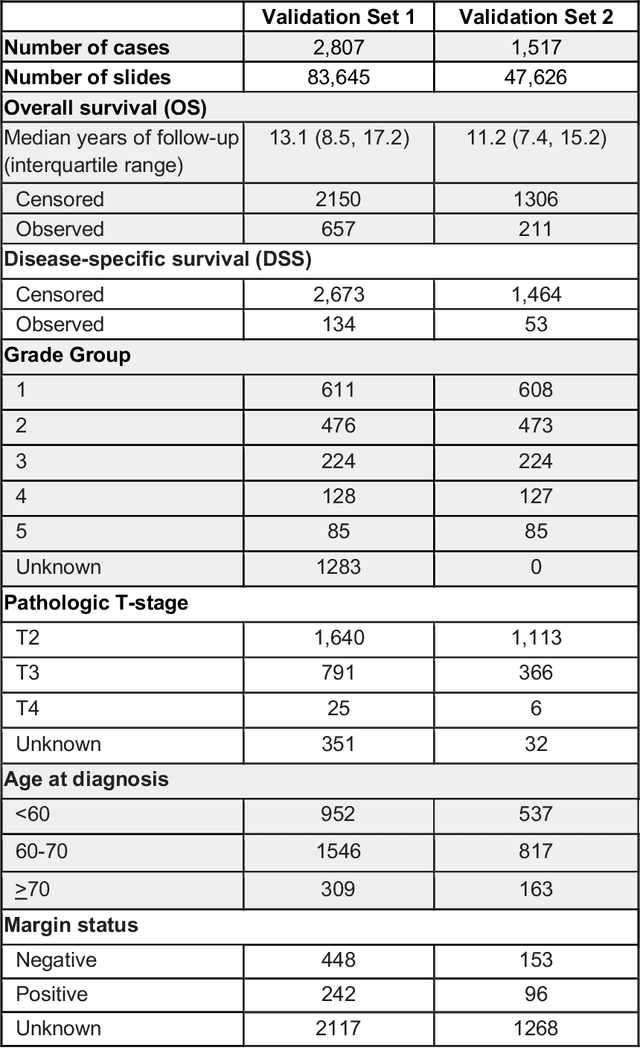
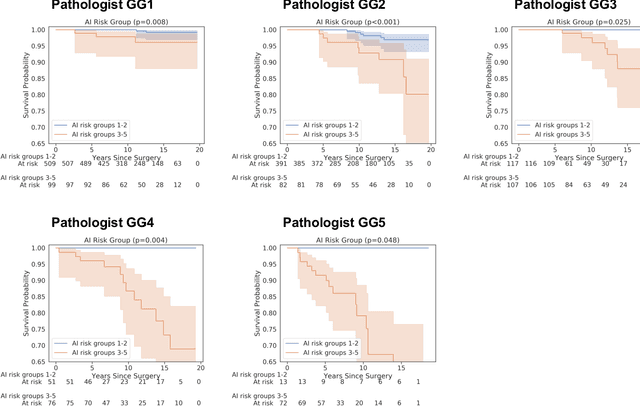
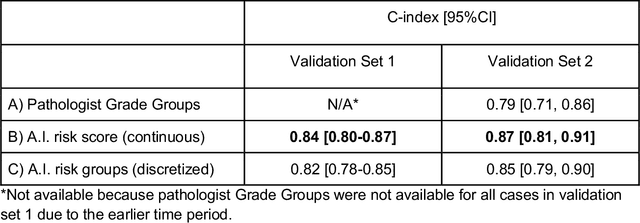
Abstract:Gleason grading of prostate cancer is an important prognostic factor but suffers from poor reproducibility, particularly among non-subspecialist pathologists. Although artificial intelligence (A.I.) tools have demonstrated Gleason grading on-par with expert pathologists, it remains an open question whether A.I. grading translates to better prognostication. In this study, we developed a system to predict prostate-cancer specific mortality via A.I.-based Gleason grading and subsequently evaluated its ability to risk-stratify patients on an independent retrospective cohort of 2,807 prostatectomy cases from a single European center with 5-25 years of follow-up (median: 13, interquartile range 9-17). The A.I.'s risk scores produced a C-index of 0.84 (95%CI 0.80-0.87) for prostate cancer-specific mortality. Upon discretizing these risk scores into risk groups analogous to pathologist Grade Groups (GG), the A.I. had a C-index of 0.82 (95%CI 0.78-0.85). On the subset of cases with a GG in the original pathology report (n=1,517), the A.I.'s C-indices were 0.87 and 0.85 for continuous and discrete grading, respectively, compared to 0.79 (95%CI 0.71-0.86) for GG obtained from the reports. These represent improvements of 0.08 (95%CI 0.01-0.15) and 0.07 (95%CI 0.00-0.14) respectively. Our results suggest that A.I.-based Gleason grading can lead to effective risk-stratification and warrants further evaluation for improving disease management.
Interpretable Survival Prediction for Colorectal Cancer using Deep Learning
Nov 17, 2020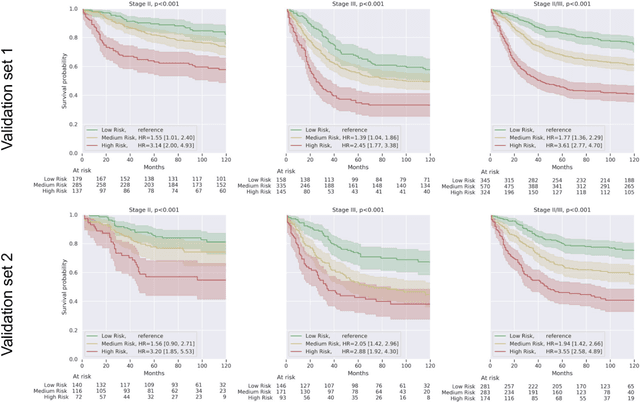
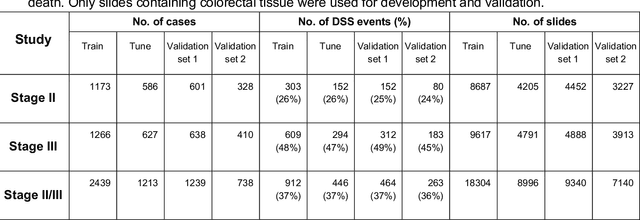
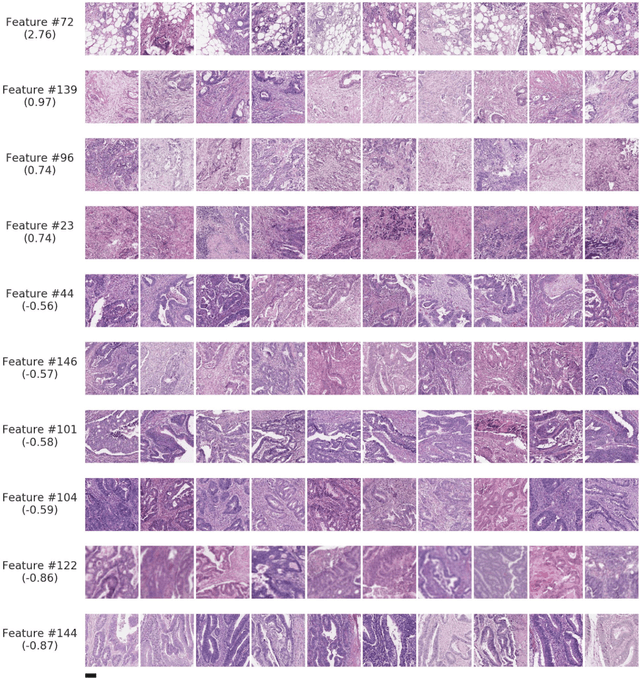
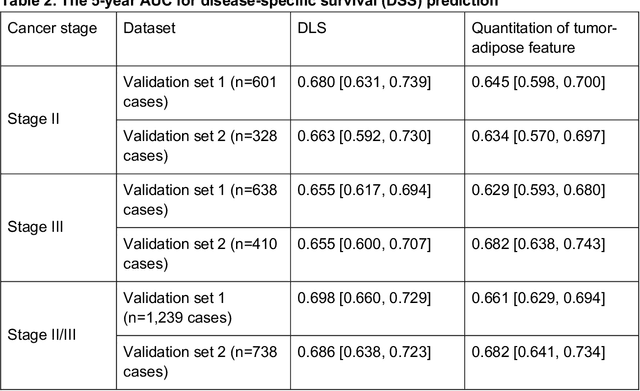
Abstract:Deriving interpretable prognostic features from deep-learning-based prognostic histopathology models remains a challenge. In this study, we developed a deep learning system (DLS) for predicting disease specific survival for stage II and III colorectal cancer using 3,652 cases (27,300 slides). When evaluated on two validation datasets containing 1,239 cases (9,340 slides) and 738 cases (7,140 slides) respectively, the DLS achieved a 5-year disease-specific survival AUC of 0.70 (95%CI 0.66-0.73) and 0.69 (95%CI 0.64-0.72), and added significant predictive value to a set of 9 clinicopathologic features. To interpret the DLS, we explored the ability of different human-interpretable features to explain the variance in DLS scores. We observed that clinicopathologic features such as T-category, N-category, and grade explained a small fraction of the variance in DLS scores (R2=18% in both validation sets). Next, we generated human-interpretable histologic features by clustering embeddings from a deep-learning based image-similarity model and showed that they explain the majority of the variance (R2 of 73% to 80%). Furthermore, the clustering-derived feature most strongly associated with high DLS scores was also highly prognostic in isolation. With a distinct visual appearance (poorly differentiated tumor cell clusters adjacent to adipose tissue), this feature was identified by annotators with 87.0-95.5% accuracy. Our approach can be used to explain predictions from a prognostic deep learning model and uncover potentially-novel prognostic features that can be reliably identified by people for future validation studies.
A deep learning system for differential diagnosis of skin diseases
Sep 11, 2019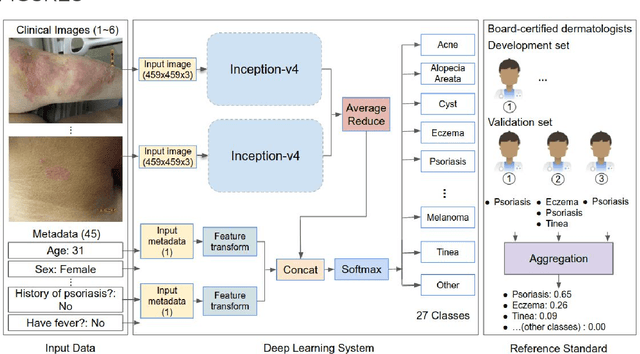
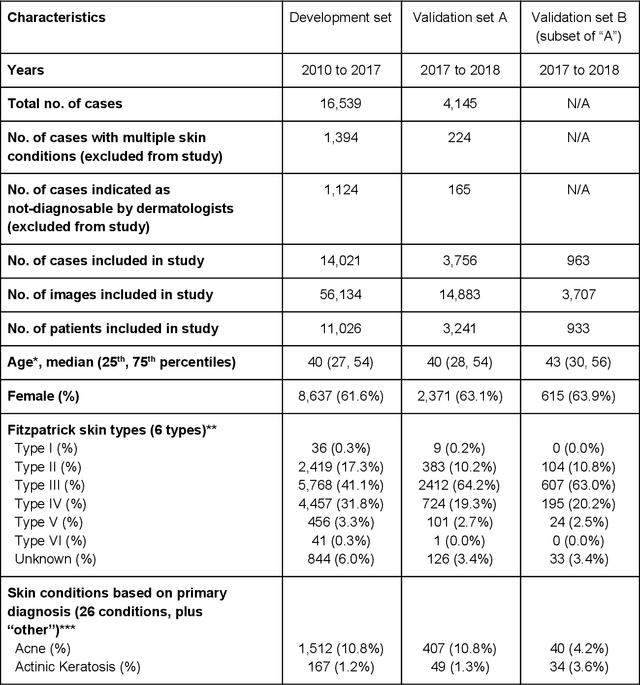
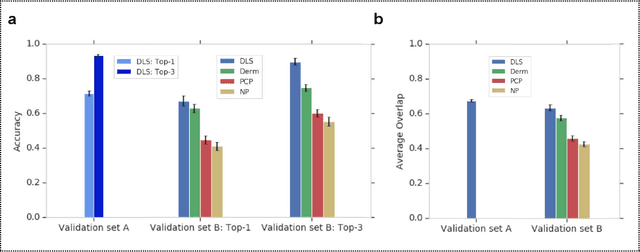
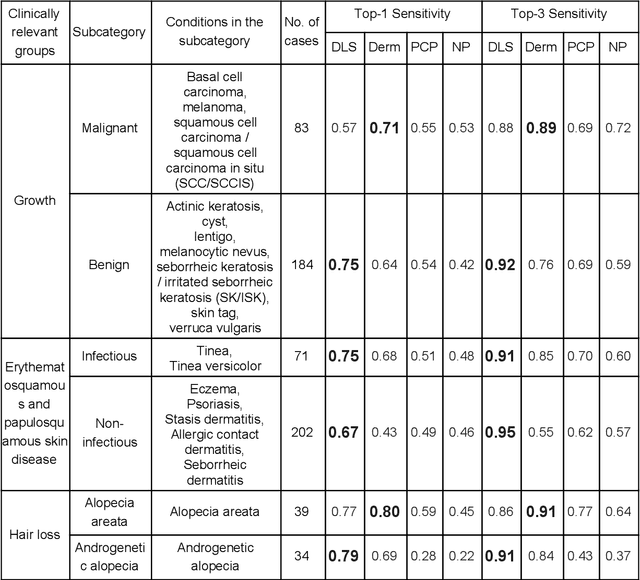
Abstract:Skin conditions affect an estimated 1.9 billion people worldwide. A shortage of dermatologists causes long wait times and leads patients to seek dermatologic care from general practitioners. However, the diagnostic accuracy of general practitioners has been reported to be only 0.24-0.70 (compared to 0.77-0.96 for dermatologists), resulting in referral errors, delays in care, and errors in diagnosis and treatment. In this paper, we developed a deep learning system (DLS) to provide a differential diagnosis of skin conditions for clinical cases (skin photographs and associated medical histories). The DLS distinguishes between 26 skin conditions that represent roughly 80% of the volume of skin conditions seen in primary care. The DLS was developed and validated using de-identified cases from a teledermatology practice serving 17 clinical sites via a temporal split: the first 14,021 cases for development and the last 3,756 cases for validation. On the validation set, where a panel of three board-certified dermatologists defined the reference standard for every case, the DLS achieved 0.71 and 0.93 top-1 and top-3 accuracies respectively. For a random subset of the validation set (n=963 cases), 18 clinicians reviewed the cases for comparison. On this subset, the DLS achieved a 0.67 top-1 accuracy, non-inferior to board-certified dermatologists (0.63, p<0.001), and higher than primary care physicians (PCPs, 0.45) and nurse practitioners (NPs, 0.41). The top-3 accuracy showed a similar trend: 0.90 DLS, 0.75 dermatologists, 0.60 PCPs, and 0.55 NPs. These results highlight the potential of the DLS to augment general practitioners to accurately diagnose skin conditions by suggesting differential diagnoses that may not have been considered. Future work will be needed to prospectively assess the clinical impact of using this tool in actual clinical workflows.
Similar Image Search for Histopathology: SMILY
Feb 06, 2019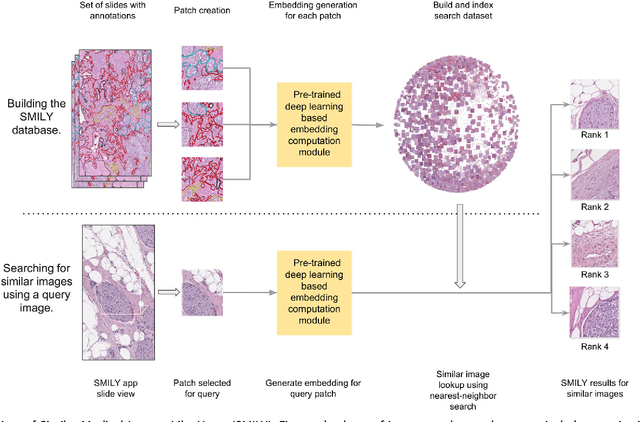
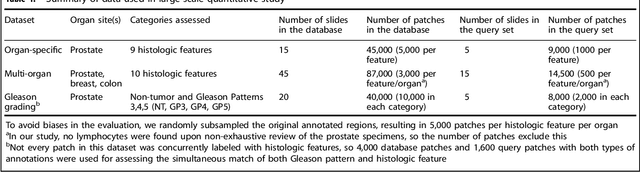
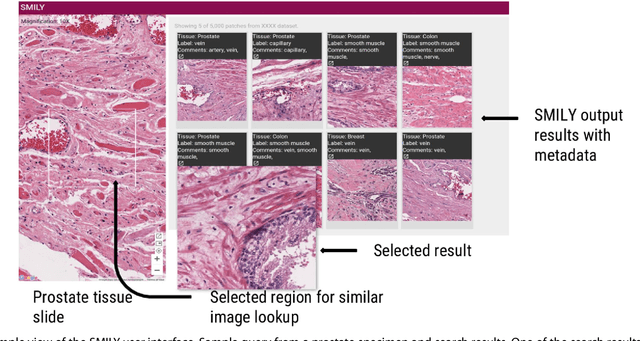
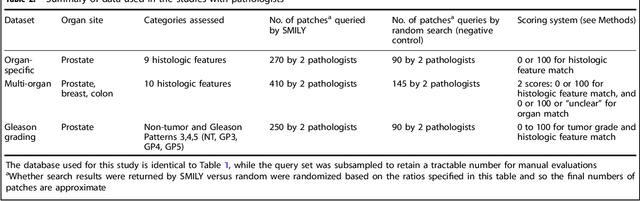
Abstract:The increasing availability of large institutional and public histopathology image datasets is enabling the searching of these datasets for diagnosis, research, and education. Though these datasets typically have associated metadata such as diagnosis or clinical notes, even carefully curated datasets rarely contain annotations of the location of regions of interest on each image. Because pathology images are extremely large (up to 100,000 pixels in each dimension), further laborious visual search of each image may be needed to find the feature of interest. In this paper, we introduce a deep learning based reverse image search tool for histopathology images: Similar Medical Images Like Yours (SMILY). We assessed SMILY's ability to retrieve search results in two ways: using pathologist-provided annotations, and via prospective studies where pathologists evaluated the quality of SMILY search results. As a negative control in the second evaluation, pathologists were blinded to whether search results were retrieved by SMILY or randomly. In both types of assessments, SMILY was able to retrieve search results with similar histologic features, organ site, and prostate cancer Gleason grade compared with the original query. SMILY may be a useful general-purpose tool in the pathologist's arsenal, to improve the efficiency of searching large archives of histopathology images, without the need to develop and implement specific tools for each application.
Development and Validation of a Deep Learning Algorithm for Improving Gleason Scoring of Prostate Cancer
Nov 15, 2018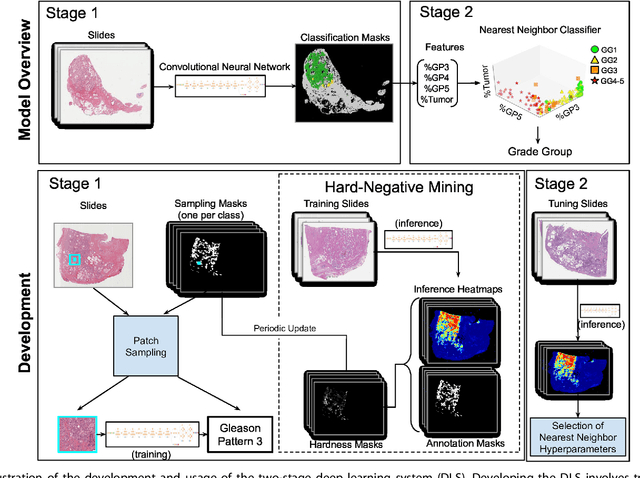
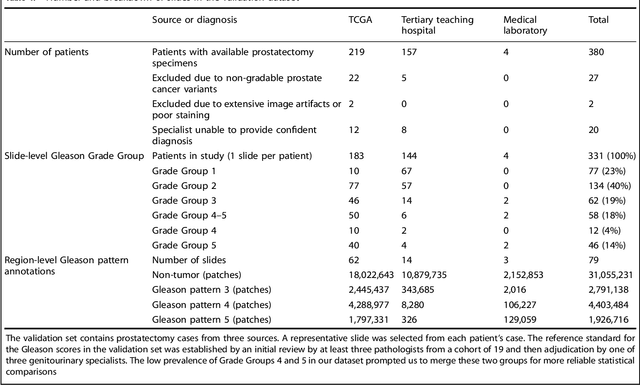
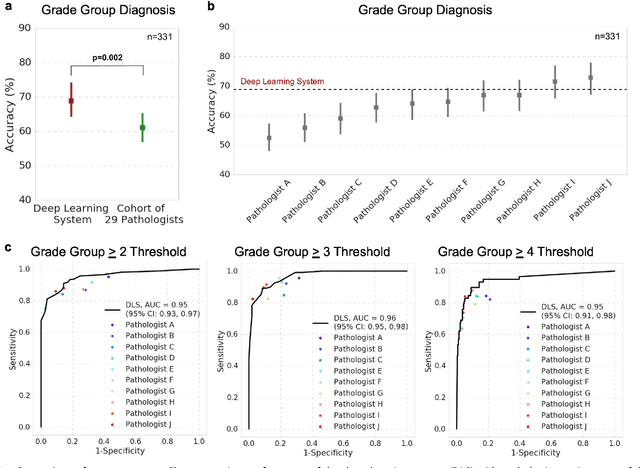
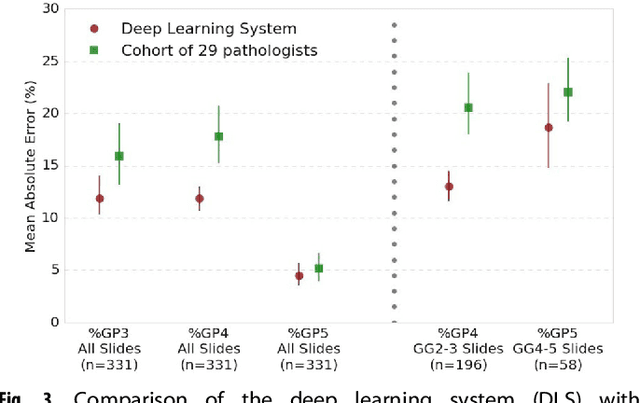
Abstract:For prostate cancer patients, the Gleason score is one of the most important prognostic factors, potentially determining treatment independent of the stage. However, Gleason scoring is based on subjective microscopic examination of tumor morphology and suffers from poor reproducibility. Here we present a deep learning system (DLS) for Gleason scoring whole-slide images of prostatectomies. Our system was developed using 112 million pathologist-annotated image patches from 1,226 slides, and evaluated on an independent validation dataset of 331 slides, where the reference standard was established by genitourinary specialist pathologists. On the validation dataset, the mean accuracy among 29 general pathologists was 0.61. The DLS achieved a significantly higher diagnostic accuracy of 0.70 (p=0.002) and trended towards better patient risk stratification in correlations to clinical follow-up data. Our approach could improve the accuracy of Gleason scoring and subsequent therapy decisions, particularly where specialist expertise is unavailable. The DLS also goes beyond the current Gleason system to more finely characterize and quantitate tumor morphology, providing opportunities for refinement of the Gleason system itself.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge