Katarzyna Bozek
ActiTect: A Generalizable Machine Learning Pipeline for REM Sleep Behavior Disorder Screening through Standardized Actigraphy
Nov 12, 2025Abstract:Isolated rapid eye movement sleep behavior disorder (iRBD) is a major prodromal marker of $α$-synucleinopathies, often preceding the clinical onset of Parkinson's disease, dementia with Lewy bodies, or multiple system atrophy. While wrist-worn actimeters hold significant potential for detecting RBD in large-scale screening efforts by capturing abnormal nocturnal movements, they become inoperable without a reliable and efficient analysis pipeline. This study presents ActiTect, a fully automated, open-source machine learning tool to identify RBD from actigraphy recordings. To ensure generalizability across heterogeneous acquisition settings, our pipeline includes robust preprocessing and automated sleep-wake detection to harmonize multi-device data and extract physiologically interpretable motion features characterizing activity patterns. Model development was conducted on a cohort of 78 individuals, yielding strong discrimination under nested cross-validation (AUROC = 0.95). Generalization was confirmed on a blinded local test set (n = 31, AUROC = 0.86) and on two independent external cohorts (n = 113, AUROC = 0.84; n = 57, AUROC = 0.94). To assess real-world robustness, leave-one-dataset-out cross-validation across the internal and external cohorts demonstrated consistent performance (AUROC range = 0.84-0.89). A complementary stability analysis showed that key predictive features remained reproducible across datasets, supporting the final pooled multi-center model as a robust pre-trained resource for broader deployment. By being open-source and easy to use, our tool promotes widespread adoption and facilitates independent validation and collaborative improvements, thereby advancing the field toward a unified and generalizable RBD detection model using wearable devices.
Histo-Miner: Deep Learning based Tissue Features Extraction Pipeline from H&E Whole Slide Images of Cutaneous Squamous Cell Carcinoma
May 07, 2025Abstract:Recent advancements in digital pathology have enabled comprehensive analysis of Whole-Slide Images (WSI) from tissue samples, leveraging high-resolution microscopy and computational capabilities. Despite this progress, there is a lack of labeled datasets and open source pipelines specifically tailored for analysis of skin tissue. Here we propose Histo-Miner, a deep learning-based pipeline for analysis of skin WSIs and generate two datasets with labeled nuclei and tumor regions. We develop our pipeline for the analysis of patient samples of cutaneous squamous cell carcinoma (cSCC), a frequent non-melanoma skin cancer. Utilizing the two datasets, comprising 47,392 annotated cell nuclei and 144 tumor-segmented WSIs respectively, both from cSCC patients, Histo-Miner employs convolutional neural networks and vision transformers for nucleus segmentation and classification as well as tumor region segmentation. Performance of trained models positively compares to state of the art with multi-class Panoptic Quality (mPQ) of 0.569 for nucleus segmentation, macro-averaged F1 of 0.832 for nucleus classification and mean Intersection over Union (mIoU) of 0.884 for tumor region segmentation. From these predictions we generate a compact feature vector summarizing tissue morphology and cellular interactions, which can be used for various downstream tasks. Here, we use Histo-Miner to predict cSCC patient response to immunotherapy based on pre-treatment WSIs from 45 patients. Histo-Miner identifies percentages of lymphocytes, the granulocyte to lymphocyte ratio in tumor vicinity and the distances between granulocytes and plasma cells in tumors as predictive features for therapy response. This highlights the applicability of Histo-Miner to clinically relevant scenarios, providing direct interpretation of the classification and insights into the underlying biology.
KPIs 2024 Challenge: Advancing Glomerular Segmentation from Patch- to Slide-Level
Feb 11, 2025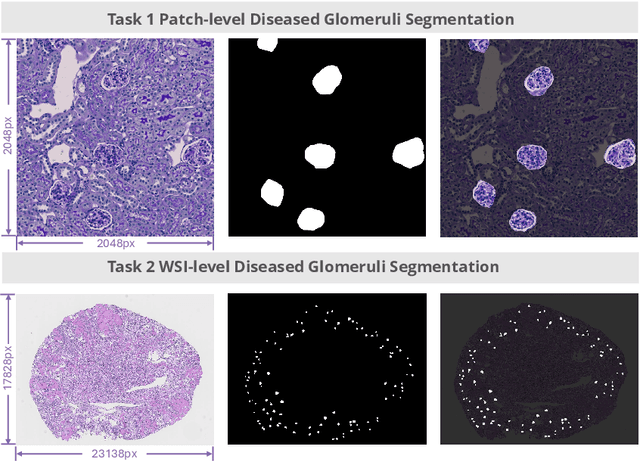
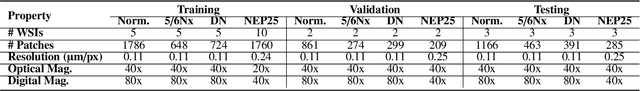
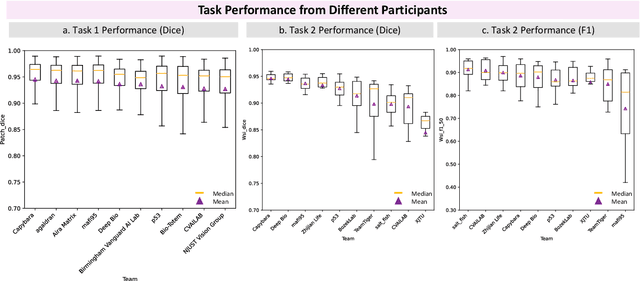

Abstract:Chronic kidney disease (CKD) is a major global health issue, affecting over 10% of the population and causing significant mortality. While kidney biopsy remains the gold standard for CKD diagnosis and treatment, the lack of comprehensive benchmarks for kidney pathology segmentation hinders progress in the field. To address this, we organized the Kidney Pathology Image Segmentation (KPIs) Challenge, introducing a dataset that incorporates preclinical rodent models of CKD with over 10,000 annotated glomeruli from 60+ Periodic Acid Schiff (PAS)-stained whole slide images. The challenge includes two tasks, patch-level segmentation and whole slide image segmentation and detection, evaluated using the Dice Similarity Coefficient (DSC) and F1-score. By encouraging innovative segmentation methods that adapt to diverse CKD models and tissue conditions, the KPIs Challenge aims to advance kidney pathology analysis, establish new benchmarks, and enable precise, large-scale quantification for disease research and diagnosis.
Fine-tuning a Multiple Instance Learning Feature Extractor with Masked Context Modelling and Knowledge Distillation
Mar 08, 2024



Abstract:The first step in Multiple Instance Learning (MIL) algorithms for Whole Slide Image (WSI) classification consists of tiling the input image into smaller patches and computing their feature vectors produced by a pre-trained feature extractor model. Feature extractor models that were pre-trained with supervision on ImageNet have proven to transfer well to this domain, however, this pre-training task does not take into account that visual information in neighboring patches is highly correlated. Based on this observation, we propose to increase downstream MIL classification by fine-tuning the feature extractor model using \textit{Masked Context Modelling with Knowledge Distillation}. In this task, the feature extractor model is fine-tuned by predicting masked patches in a bigger context window. Since reconstructing the input image would require a powerful image generation model, and our goal is not to generate realistically looking image patches, we predict instead the feature vectors produced by a larger teacher network. A single epoch of the proposed task suffices to increase the downstream performance of the feature-extractor model when used in a MIL scenario, even capable of outperforming the downstream performance of the teacher model, while being considerably smaller and requiring a fraction of its compute.
Language models are good pathologists: using attention-based sequence reduction and text-pretrained transformers for efficient WSI classification
Nov 14, 2022



Abstract:In digital pathology, Whole Slide Image (WSI) analysis is usually formulated as a Multiple Instance Learning (MIL) problem. Although transformer-based architectures have been used for WSI classification, these methods require modifications to adapt them to specific challenges of this type of image data. Despite their power across domains, reference transformer models in classical Computer Vision (CV) and Natural Language Processing (NLP) tasks are not used for pathology slide analysis. In this work we demonstrate the use of standard, frozen, text-pretrained, transformer language models in application to WSI classification. We propose SeqShort, a multi-head attention-based sequence reduction input layer to summarize each WSI in a fixed and short size sequence of instances. This allows us to reduce the computational costs of self-attention on long sequences, and to include positional information that is unavailable in other MIL approaches. We demonstrate the effectiveness of our methods in the task of cancer subtype classification, without the need of designing a WSI-specific transformer or performing in-domain self-supervised pretraining, while keeping a reduced compute budget and number of trainable parameters.
Pixel personality for dense object tracking in a 2D honeybee hive
Dec 31, 2018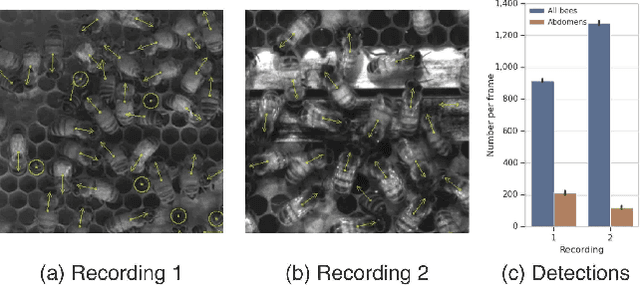
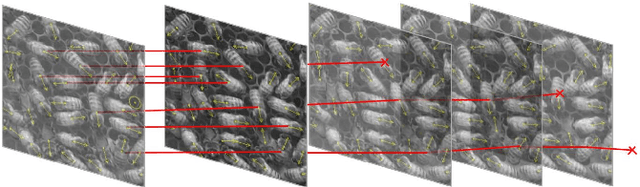
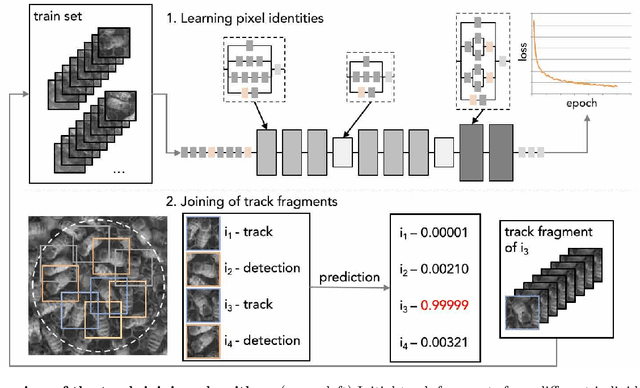
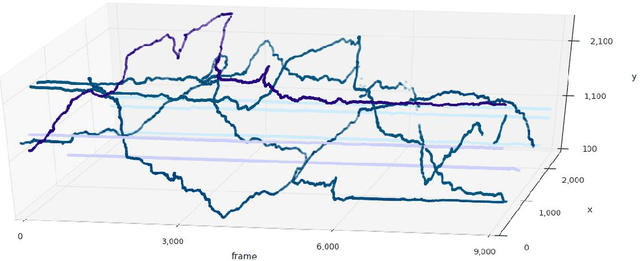
Abstract:Tracking large numbers of densely-arranged, interacting objects is challenging due to occlusions and the resulting complexity of possible trajectory combinations, as well as the sparsity of relevant, labeled datasets. Here we describe a novel technique of collective tracking in the model environment of a 2D honeybee hive in which sample colonies consist of $N\sim10^3$ highly similar individuals, tightly packed, and in rapid, irregular motion. Such a system offers universal challenges for multi-object tracking, while being conveniently accessible for image recording. We first apply an accurate, segmentation-based object detection method to build initial short trajectory segments by matching object configurations based on class, position and orientation. We then join these tracks into full single object trajectories by creating an object recognition model which is adaptively trained to recognize honeybee individuals through their visual appearance across multiple frames, an attribute we denote as pixel personality. Overall, we reconstruct ~46% of the trajectories in 5 min recordings from two different hives and over 71% of the tracks for at least 2 min. We provide validated trajectories spanning 3000 video frames of 876 unmarked moving bees in two distinct colonies in different locations and filmed with different pixel resolutions, which we expect to be useful in the further development of general-purpose tracking solutions.
Towards dense object tracking in a 2D honeybee hive
Dec 22, 2017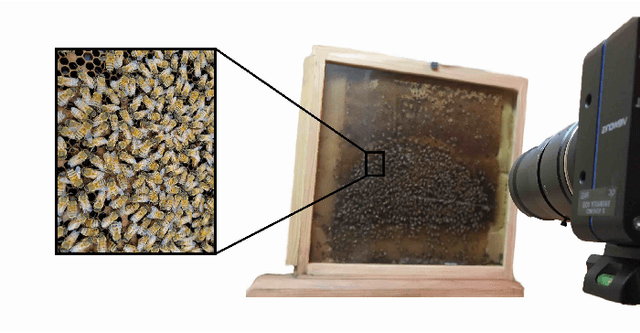
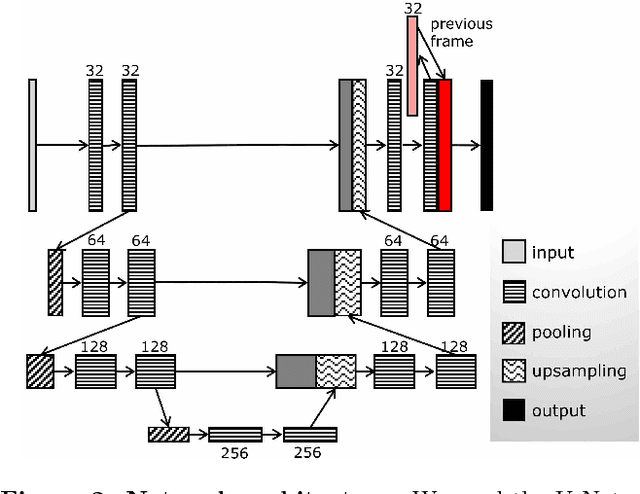
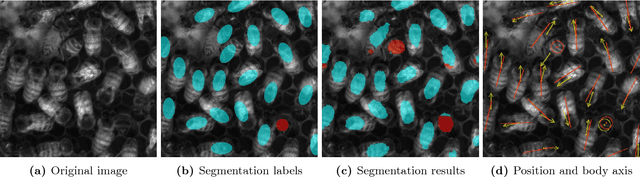
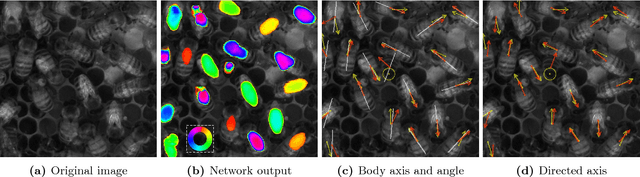
Abstract:From human crowds to cells in tissue, the detection and efficient tracking of multiple objects in dense configurations is an important and unsolved problem. In the past, limitations of image analysis have restricted studies of dense groups to tracking a single or subset of marked individuals, or to coarse-grained group-level dynamics, all of which yield incomplete information. Here, we combine convolutional neural networks (CNNs) with the model environment of a honeybee hive to automatically recognize all individuals in a dense group from raw image data. We create new, adapted individual labeling and use the segmentation architecture U-Net with a loss function dependent on both object identity and orientation. We additionally exploit temporal regularities of the video recording in a recurrent manner and achieve near human-level performance while reducing the network size by 94% compared to the original U-Net architecture. Given our novel application of CNNs, we generate extensive problem-specific image data in which labeled examples are produced through a custom interface with Amazon Mechanical Turk. This dataset contains over 375,000 labeled bee instances across 720 video frames at 2 FPS, representing an extensive resource for the development and testing of tracking methods. We correctly detect 96% of individuals with a location error of ~7% of a typical body dimension, and orientation error of 12 degrees, approximating the variability of human raters. Our results provide an important step towards efficient image-based dense object tracking by allowing for the accurate determination of object location and orientation across time-series image data efficiently within one network architecture.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge