Justin Sybrandt
Literature-based Discovery for Landscape Planning
Jun 05, 2023Abstract:This project demonstrates how medical corpus hypothesis generation, a knowledge discovery field of AI, can be used to derive new research angles for landscape and urban planners. The hypothesis generation approach herein consists of a combination of deep learning with topic modeling, a probabilistic approach to natural language analysis that scans aggregated research databases for words that can be grouped together based on their subject matter commonalities; the word groups accordingly form topics that can provide implicit connections between two general research terms. The hypothesis generation system AGATHA was used to identify likely conceptual relationships between emerging infectious diseases (EIDs) and deforestation, with the objective of providing landscape planners guidelines for productive research directions to help them formulate research hypotheses centered on deforestation and EIDs that will contribute to the broader health field that asserts causal roles of landscape-level issues. This research also serves as a partial proof-of-concept for the application of medical database hypothesis generation to medicine-adjacent hypothesis discovery.
SmartChoices: Augmenting Software with Learned Implementations
Apr 12, 2023



Abstract:We are living in a golden age of machine learning. Powerful models are being trained to perform many tasks far better than is possible using traditional software engineering approaches alone. However, developing and deploying those models in existing software systems remains difficult. In this paper we present SmartChoices, a novel approach to incorporating machine learning into mature software stacks easily, safely, and effectively. We explain the overall design philosophy and present case studies using SmartChoices within large scale industrial systems.
Accelerating COVID-19 research with graph mining and transformer-based learning
Feb 10, 2021Abstract:In 2020, the White House released the, "Call to Action to the Tech Community on New Machine Readable COVID-19 Dataset," wherein artificial intelligence experts are asked to collect data and develop text mining techniques that can help the science community answer high-priority scientific questions related to COVID-19. The Allen Institute for AI and collaborators announced the availability of a rapidly growing open dataset of publications, the COVID-19 Open Research Dataset (CORD-19). As the pace of research accelerates, biomedical scientists struggle to stay current. To expedite their investigations, scientists leverage hypothesis generation systems, which can automatically inspect published papers to discover novel implicit connections. We present an automated general purpose hypothesis generation systems AGATHA-C and AGATHA-GP for COVID-19 research. The systems are based on graph-mining and the transformer model. The systems are massively validated using retrospective information rediscovery and proactive analysis involving human-in-the-loop expert analysis. Both systems achieve high-quality predictions across domains (in some domains up to 0.97% ROC AUC) in fast computational time and are released to the broad scientific community to accelerate biomedical research. In addition, by performing the domain expert curated study, we show that the systems are able to discover on-going research findings such as the relationship between COVID-19 and oxytocin hormone.
Accelerating Text Mining Using Domain-Specific Stop Word Lists
Nov 18, 2020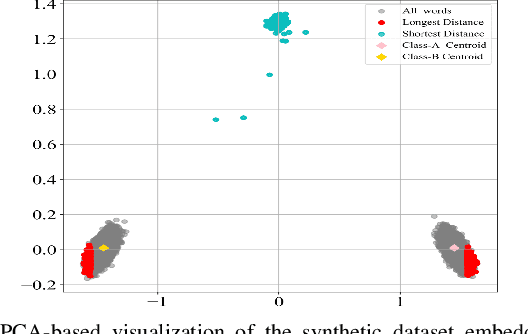
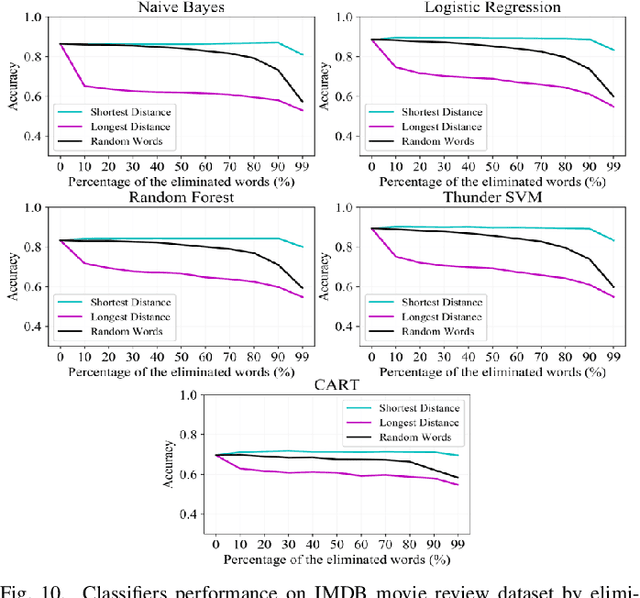
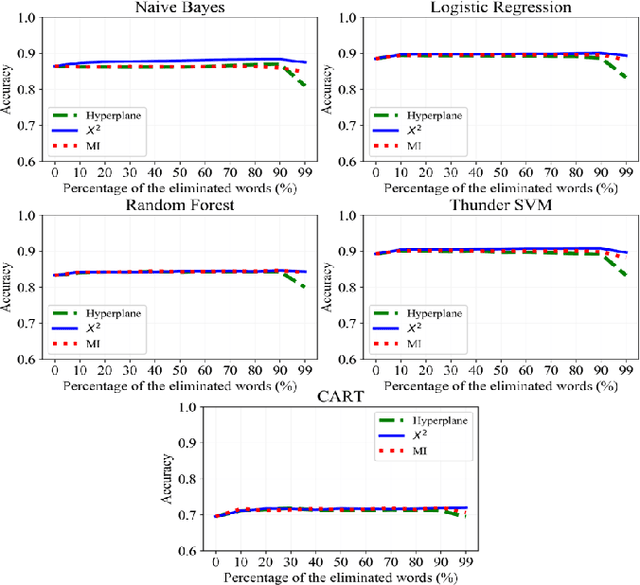
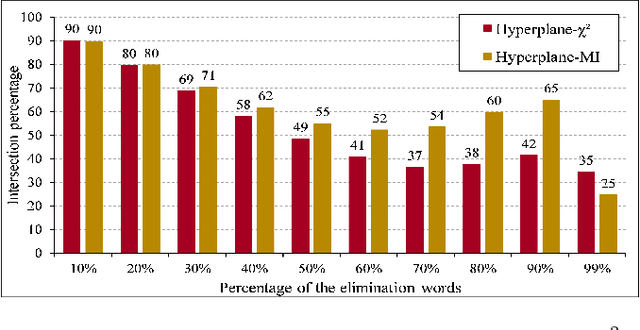
Abstract:Text preprocessing is an essential step in text mining. Removing words that can negatively impact the quality of prediction algorithms or are not informative enough is a crucial storage-saving technique in text indexing and results in improved computational efficiency. Typically, a generic stop word list is applied to a dataset regardless of the domain. However, many common words are different from one domain to another but have no significance within a particular domain. Eliminating domain-specific common words in a corpus reduces the dimensionality of the feature space, and improves the performance of text mining tasks. In this paper, we present a novel mathematical approach for the automatic extraction of domain-specific words called the hyperplane-based approach. This new approach depends on the notion of low dimensional representation of the word in vector space and its distance from hyperplane. The hyperplane-based approach can significantly reduce text dimensionality by eliminating irrelevant features. We compare the hyperplane-based approach with other feature selection methods, namely \c{hi}2 and mutual information. An experimental study is performed on three different datasets and five classification algorithms, and measure the dimensionality reduction and the increase in the classification performance. Results indicate that the hyperplane-based approach can reduce the dimensionality of the corpus by 90% and outperforms mutual information. The computational time to identify the domain-specific words is significantly lower than mutual information.
Unsupervised Hierarchical Graph Representation Learning by Mutual Information Maximization
Apr 02, 2020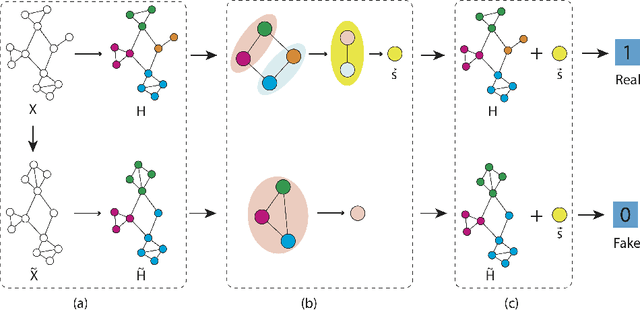
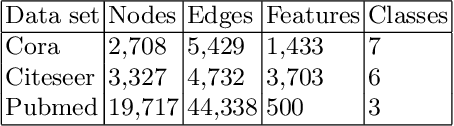
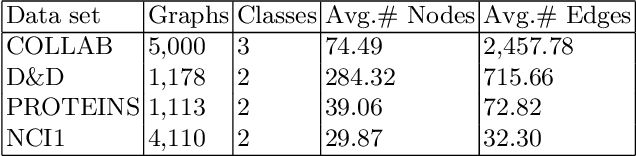
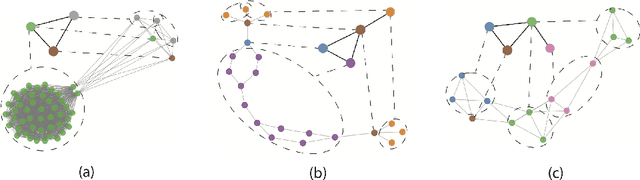
Abstract:Graph representation learning based on graph neural networks (GNNs) can greatly improve the performance of downstream tasks, such as node and graph classification. However, the general GNN models do not aggregate node information in a hierarchical manner, and can miss key higher-order structural features of many graphs. The hierarchical aggregation also enables the graph representations to be explainable. In addition, supervised graph representation learning requires labeled data, which is expensive and error-prone. To address these issues, we present an unsupervised graph representation learning method, Unsupervised Hierarchical Graph Representation (UHGR), which can generate hierarchical representations of graphs. Our method focuses on maximizing mutual information between "local" and high-level "global" representations, which enables us to learn the node embeddings and graph embeddings without any labeled data. To demonstrate the effectiveness of the proposed method, we perform the node and graph classification using the learned node and graph embeddings. The results show that the proposed method achieves comparable results to state-of-the-art supervised methods on several benchmarks. In addition, our visualization of hierarchical representations indicates that our method can capture meaningful and interpretable clusters.
CBAG: Conditional Biomedical Abstract Generation
Feb 13, 2020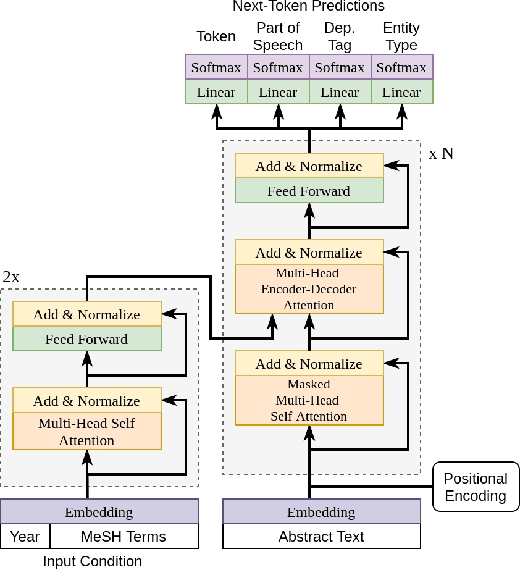
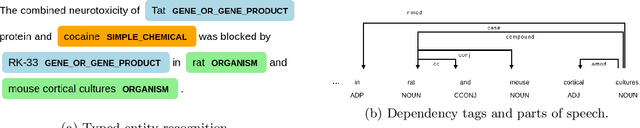
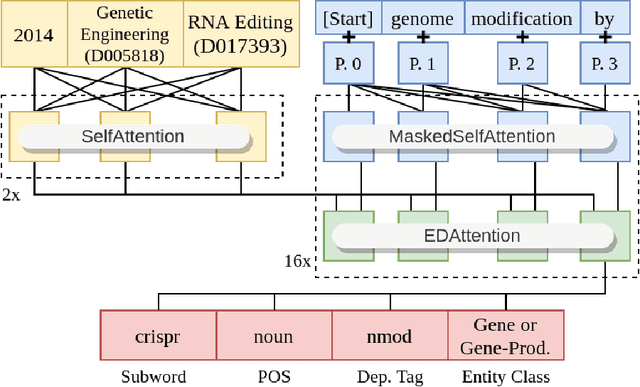
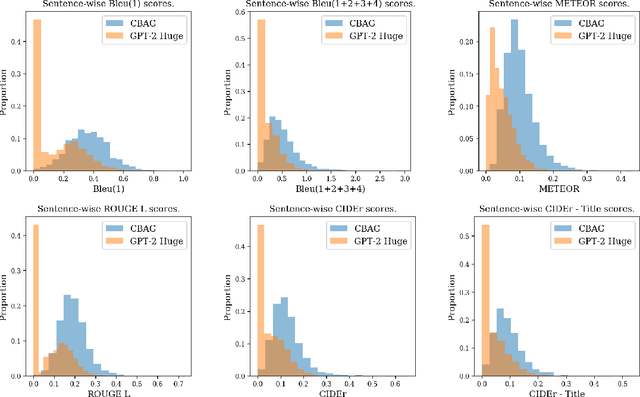
Abstract:Biomedical research papers use significantly different language and jargon when compared to typical English text, which reduces the utility of pre-trained NLP models in this domain. Meanwhile Medline, a database of biomedical abstracts, introduces nearly a million new documents per-year. Applications that could benefit from understanding this wealth of publicly available information, such as scientific writing assistants, chat-bots, or descriptive hypothesis generation systems, require new domain-centered approaches. A conditional language model, one that learns the probability of words given some a priori criteria, is a fundamental building block in many such applications. We propose a transformer-based conditional language model with a shallow encoder "condition" stack, and a deep "language model" stack of multi-headed attention blocks. The condition stack encodes metadata used to alter the output probability distribution of the language model stack. We sample this distribution in order to generate biomedical abstracts given only a proposed title, an intended publication year, and a set of keywords. Using typical natural language generation metrics, we demonstrate that this proposed approach is more capable of producing non-trivial relevant entities within the abstract body than the 1.5B parameter GPT-2 language model.
AGATHA: Automatic Graph-mining And Transformer based Hypothesis generation Approach
Feb 13, 2020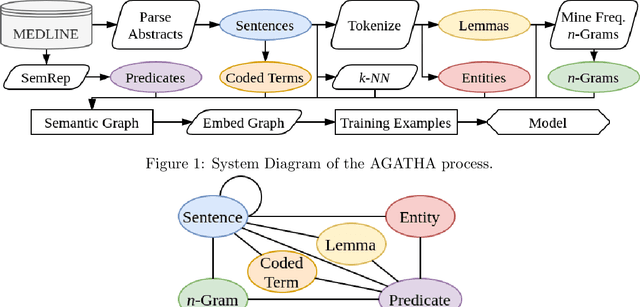
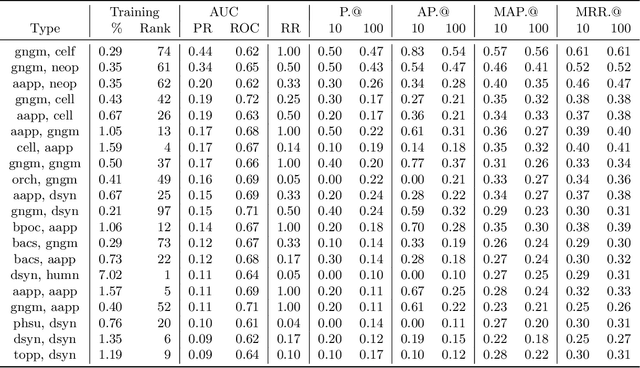
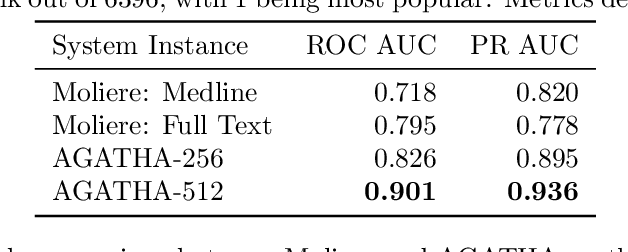
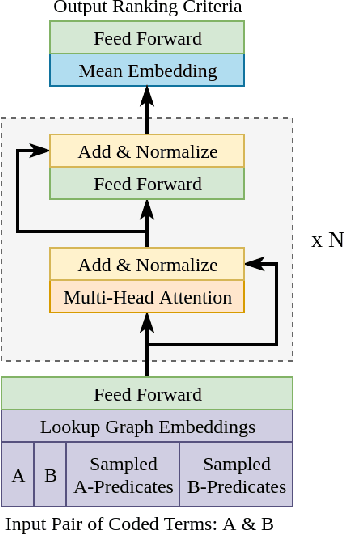
Abstract:Medical research is risky and expensive. Drug discovery, as an example, requires that researchers efficiently winnow thousands of potential targets to a small candidate set for more thorough evaluation. However, research groups spend significant time and money to perform the experiments necessary to determine this candidate set long before seeing intermediate results. Hypothesis generation systems address this challenge by mining the wealth of publicly available scientific information to predict plausible research directions. We present AGATHA, a deep-learning hypothesis generation system that can introduce data-driven insights earlier in the discovery process. Through a learned ranking criteria, this system quickly prioritizes plausible term-pairs among entity sets, allowing us to recommend new research directions. We massively validate our system with a temporal holdout wherein we predict connections first introduced after 2015 using data published beforehand. We additionally explore biomedical sub-domains, and demonstrate AGATHA's predictive capacity across the twenty most popular relationship types. This system achieves best-in-class performance on an established benchmark, and demonstrates high recommendation scores across subdomains. Reproducibility: All code, experimental data, and pre-trained models are available online: sybrandt.com/2020/agatha
Hypergraph Partitioning With Embeddings
Sep 16, 2019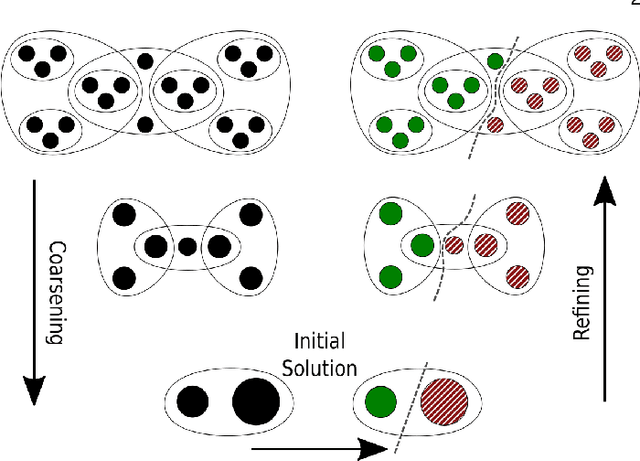
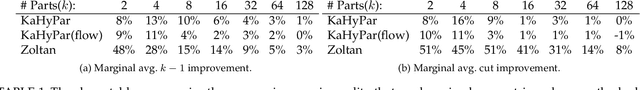
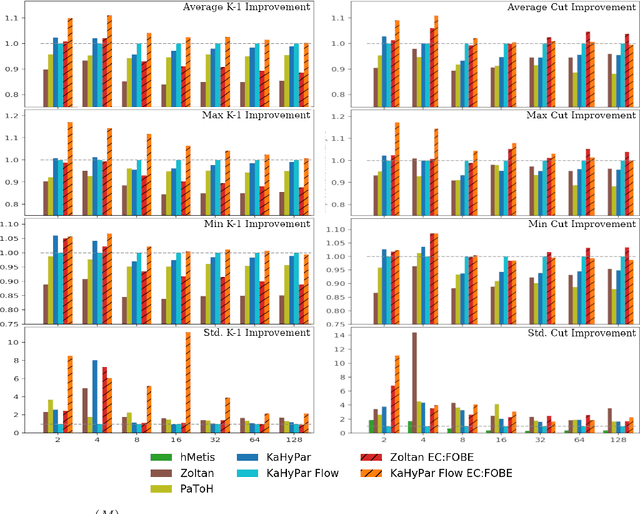
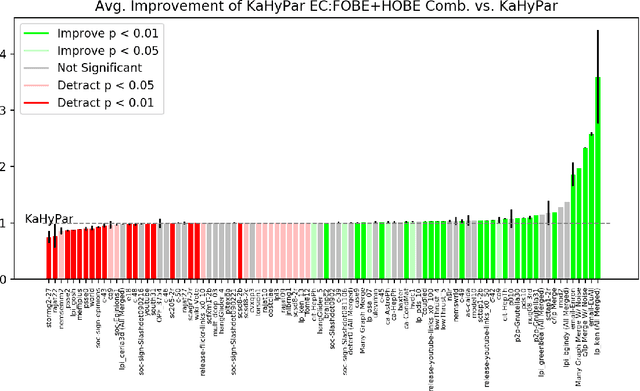
Abstract:The problem of placing circuits on a chip or distributing sparse matrix operations can be modeled as the hypergraph partitioning problem. A hypergraph is a generalization of the traditional graph wherein each "hyperedge" may connect any number of nodes. Hypergraph partitioning, therefore, is the NP-Hard problem of dividing nodes into $k$ similarly sized disjoint sets while minimizing the number of hyperedges that span multiple partitions. Due to this problem's complexity, many partitioners leverage the multilevel heuristic of iteratively "coarsening" their input to a smaller approximation until an inefficient algorithm becomes feasible. The initial solution is then propagated back to the original hypergraph, which produces a reasonably accurate result provided the coarse representation preserves structural properties of the original. The multilevel hypergraph partitioners are considered today as state-of-the-art solvers that achieve an excellent quality/running time trade-off on practical large-scale instances of different types. In order to improve the quality of multilevel hypergraph partitioners, we propose leveraging graph embeddings to better capture structural properties during the coarsening process. Our approach prioritizes dense subspaces found at the embedding, and contracts nodes according to both traditional and embedding-based similarity measures. Reproducibility: All source code, plots and experimental data are available at https://sybrandt.com/2019/partition.
FOBE and HOBE: First- and High-Order Bipartite Embeddings
May 27, 2019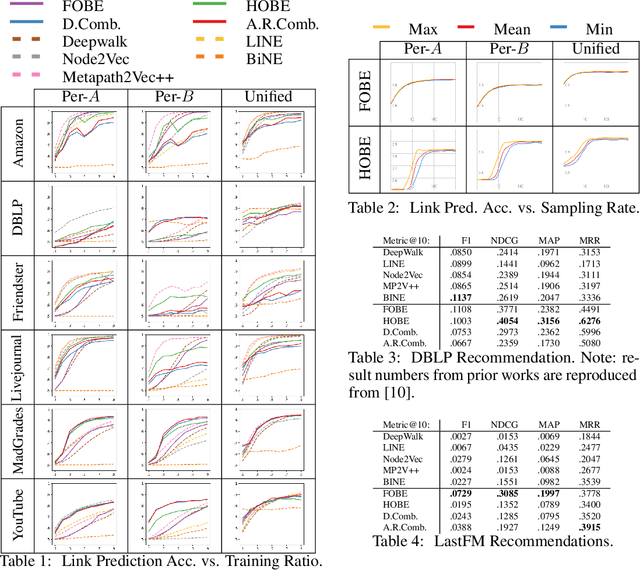
Abstract:Typical graph embeddings may not capture type-specific bipartite graph features that arise in such areas as recommender systems, data visualization, and drug discovery. Machine learning methods utilized in these applications would be better served with specialized embedding techniques. We propose two embeddings for bipartite graphs that decompose edges into sets of indirect relationships between node neighborhoods. When sampling higher-order relationships, we reinforce similarities through algebraic distance on graphs. We also introduce ensemble embeddings to combine both into a "best of both worlds" embedding. The proposed methods are evaluated on link prediction and recommendation tasks and compared with other state-of-the-art embeddings. While being all highly beneficial in applications, we demonstrate that none of the considered embeddings is clearly superior (in contrast to what is claimed in many papers), and discuss the trade offs present among them. Reproducibility: Our code, data sets, and results are all publicly available online at: http://bit.ly/fobe_hobe_code.
Large-Scale Validation of Hypothesis Generation Systems via Candidate Ranking
Oct 19, 2018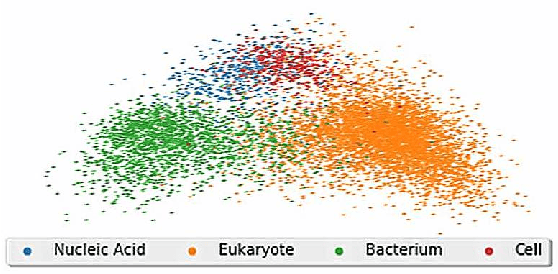
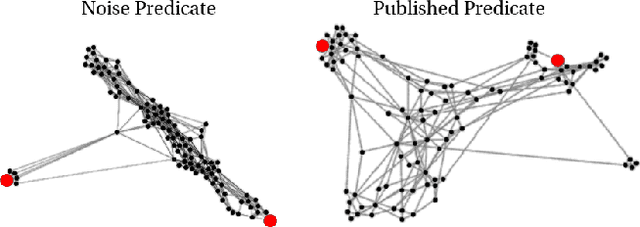
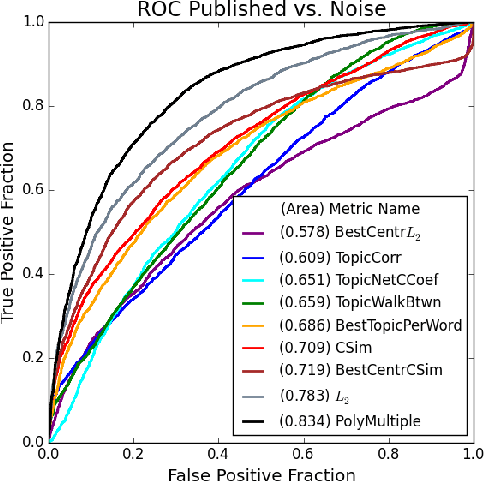
Abstract:The first step of many research projects is to define and rank a short list of candidates for study. In the modern rapidity of scientific progress, some turn to automated hypothesis generation (HG) systems to aid this process. These systems can identify implicit or overlooked connections within a large scientific corpus, and while their importance grows alongside the pace of science, they lack thorough validation. Without any standard numerical evaluation method, many validate general-purpose HG systems by rediscovering a handful of historical findings, and some wishing to be more thorough may run laboratory experiments based on automatic suggestions. These methods are expensive, time consuming, and cannot scale. Thus, we present a numerical evaluation framework for the purpose of validating HG systems that leverages thousands of validation hypotheses. This method evaluates a HG system by its ability to rank hypotheses by plausibility; a process reminiscent of human candidate selection. Because HG systems do not produce a ranking criteria, specifically those that produce topic models, we additionally present novel metrics to quantify the plausibility of hypotheses given topic model system output. Finally, we demonstrate that our proposed validation method aligns with real-world research goals by deploying our method within Moliere, our recent topic-driven HG system, in order to automatically generate a set of candidate genes related to HIV-associated neurodegenerative disease (HAND). By performing laboratory experiments based on this candidate set, we discover a new connection between HAND and Dead Box RNA Helicase 3 (DDX3). Reproducibility: code, validation data, and results can be found at sybrandt.com/2018/validation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge