Julien Cohen-Adad
HiFormer: Hierarchical Multi-scale Representations Using Transformers for Medical Image Segmentation
Jul 18, 2022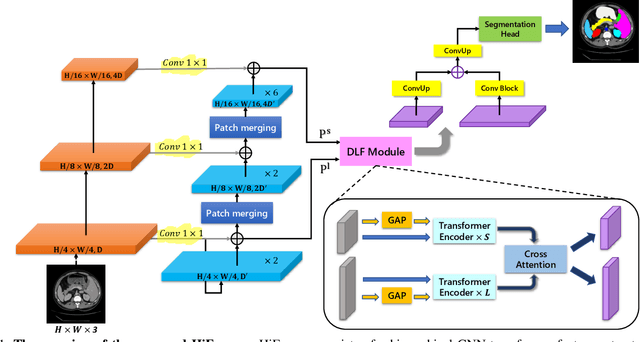
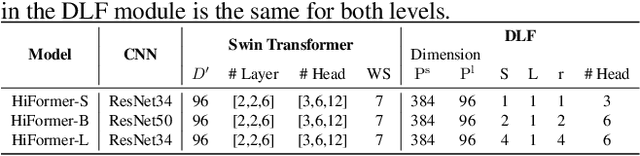
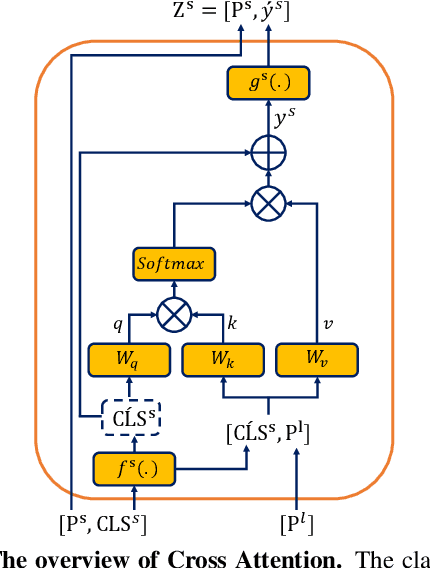
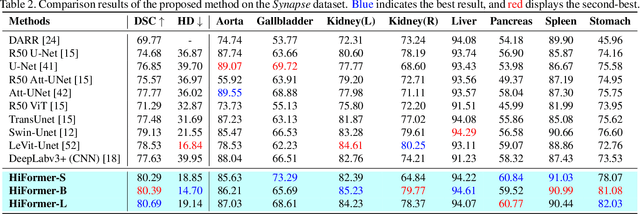
Abstract:Convolutional neural networks (CNNs) have been the consensus for medical image segmentation tasks. However, they suffer from the limitation in modeling long-range dependencies and spatial correlations due to the nature of convolution operation. Although transformers were first developed to address this issue, they fail to capture low-level features. In contrast, it is demonstrated that both local and global features are crucial for dense prediction, such as segmenting in challenging contexts. In this paper, we propose HiFormer, a novel method that efficiently bridges a CNN and a transformer for medical image segmentation. Specifically, we design two multi-scale feature representations using the seminal Swin Transformer module and a CNN-based encoder. To secure a fine fusion of global and local features obtained from the two aforementioned representations, we propose a Double-Level Fusion (DLF) module in the skip connection of the encoder-decoder structure. Extensive experiments on various medical image segmentation datasets demonstrate the effectiveness of HiFormer over other CNN-based, transformer-based, and hybrid methods in terms of computational complexity, and quantitative and qualitative results. Our code is publicly available at: https://github.com/amirhossein-kz/HiFormer
Intervertebral Disc Labeling With Learning Shape Information, A Look Once Approach
Apr 06, 2022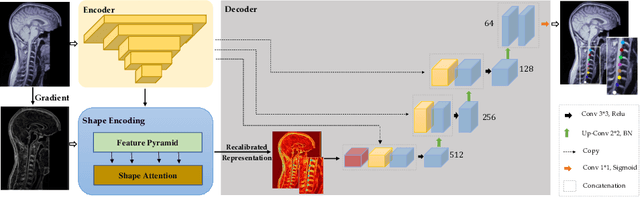
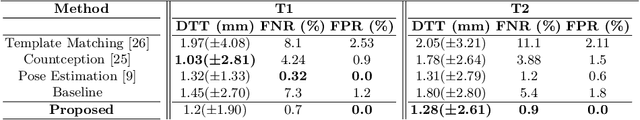
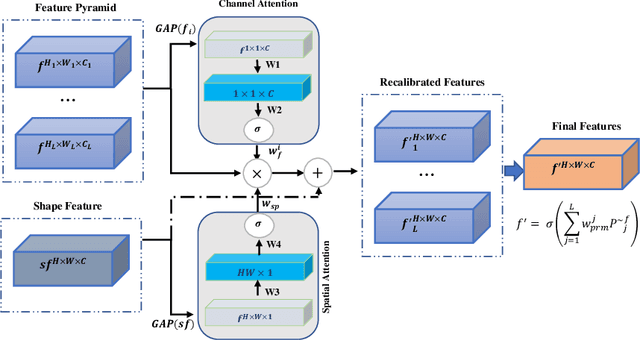
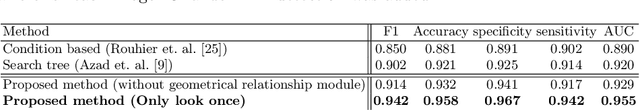
Abstract:Accurate and automatic segmentation of intervertebral discs from medical images is a critical task for the assessment of spine-related diseases such as osteoporosis, vertebral fractures, and intervertebral disc herniation. To date, various approaches have been developed in the literature which routinely relies on detecting the discs as the primary step. A disadvantage of many cohort studies is that the localization algorithm also yields false-positive detections. In this study, we aim to alleviate this problem by proposing a novel U-Net-based structure to predict a set of candidates for intervertebral disc locations. In our design, we integrate the image shape information (image gradients) to encourage the model to learn rich and generic geometrical information. This additional signal guides the model to selectively emphasize the contextual representation and suppress the less discriminative features. On the post-processing side, to further decrease the false positive rate, we propose a permutation invariant 'look once' model, which accelerates the candidate recovery procedure. In comparison with previous studies, our proposed approach does not need to perform the selection in an iterative fashion. The proposed method was evaluated on the spine generic public multi-center dataset and demonstrated superior performance compared to previous work. We have provided the implementation code in https://github.com/rezazad68/intervertebral-lookonce
Medical Image Segmentation on MRI Images with Missing Modalities: A Review
Mar 11, 2022
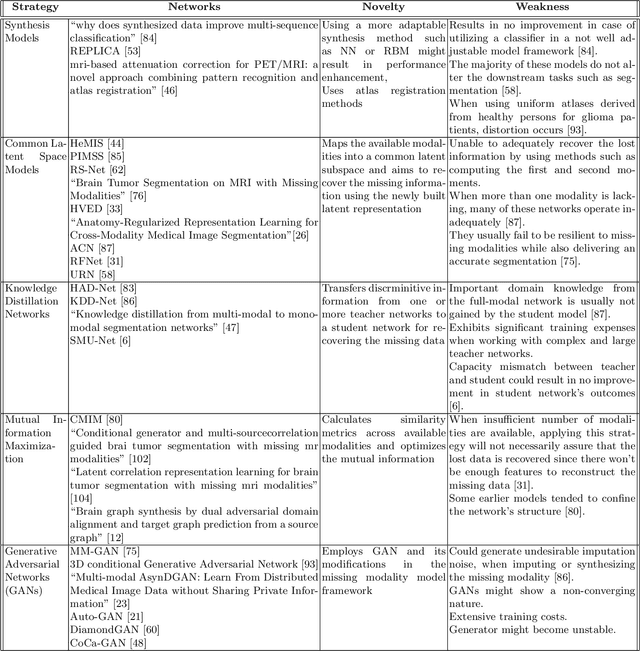
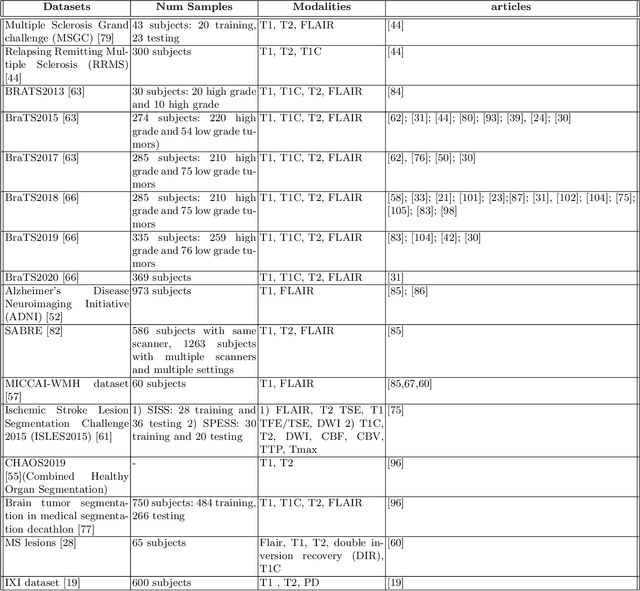
Abstract:Dealing with missing modalities in Magnetic Resonance Imaging (MRI) and overcoming their negative repercussions is considered a hurdle in biomedical imaging. The combination of a specified set of modalities, which is selected depending on the scenario and anatomical part being scanned, will provide medical practitioners with full information about the region of interest in the human body, hence the missing MRI sequences should be reimbursed. The compensation of the adverse impact of losing useful information owing to the lack of one or more modalities is a well-known challenge in the field of computer vision, particularly for medical image processing tasks including tumour segmentation, tissue classification, and image generation. Various approaches have been developed over time to mitigate this problem's negative implications and this literature review goes through a significant number of the networks that seek to do so. The approaches reviewed in this work are reviewed in detail, including earlier techniques such as synthesis methods as well as later approaches that deploy deep learning, such as common latent space models, knowledge distillation networks, mutual information maximization, and generative adversarial networks (GANs). This work discusses the most important approaches that have been offered at the time of this writing, examining the novelty, strength, and weakness of each one. Furthermore, the most commonly used MRI datasets are highlighted and described. The main goal of this research is to offer a performance evaluation of missing modality compensating networks, as well as to outline future strategies for dealing with this issue.
Label fusion and training methods for reliable representation of inter-rater uncertainty
Feb 26, 2022
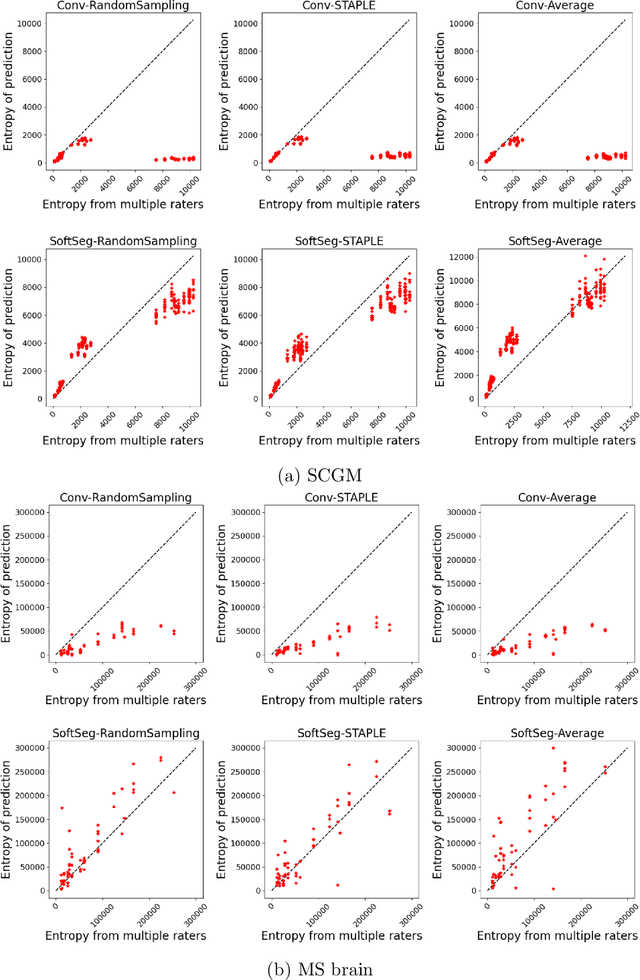
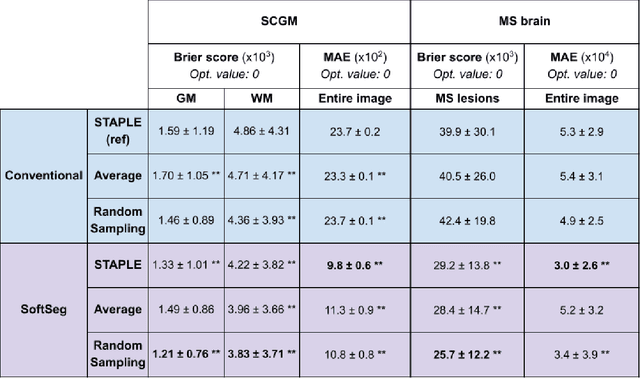
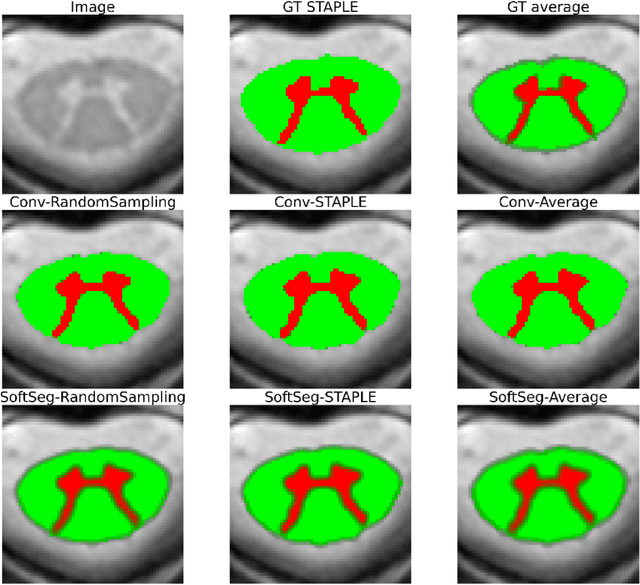
Abstract:Medical tasks are prone to inter-rater variability due to multiple factors such as image quality, professional experience and training, or guideline clarity. Training deep learning networks with annotations from multiple raters is a common practice that mitigates the model's bias towards a single expert. Reliable models generating calibrated outputs and reflecting the inter-rater disagreement are key to the integration of artificial intelligence in clinical practice. Various methods exist to take into account different expert labels. We focus on comparing three label fusion methods: STAPLE, average of the rater's segmentation, and random sampling of each rater's segmentation during training. Each label fusion method is studied using both the conventional training framework and the recently published SoftSeg framework that limits information loss by treating the segmentation task as a regression. Our results, across 10 data splittings on two public datasets, indicate that SoftSeg models, regardless of the ground truth fusion method, had better calibration and preservation of the inter-rater rater variability compared with their conventional counterparts without impacting the segmentation performance. Conventional models, i.e., trained with a Dice loss, with binary inputs, and sigmoid/softmax final activate, were overconfident and underestimated the uncertainty associated with inter-rater variability. Conversely, fusing labels by averaging with the SoftSeg framework led to underconfident outputs and overestimation of the rater disagreement. In terms of segmentation performance, the best label fusion method was different for the two datasets studied, indicating this parameter might be task-dependent. However, SoftSeg had segmentation performance systematically superior or equal to the conventionally trained models and had the best calibration and preservation of the inter-rater variability.
2D Multi-Class Model for Gray and White Matter Segmentation of the Cervical Spinal Cord at 7T
Oct 13, 2021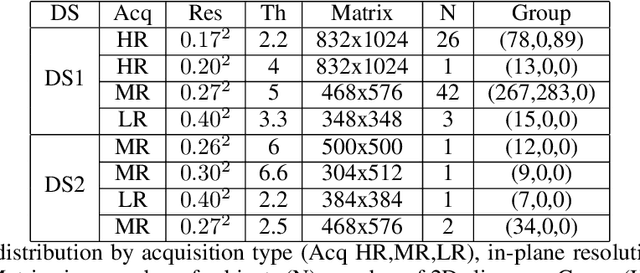
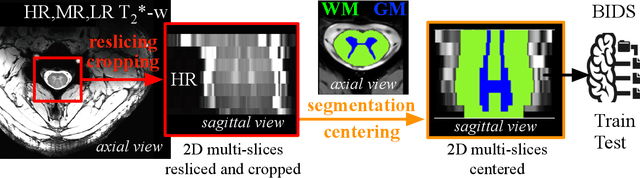
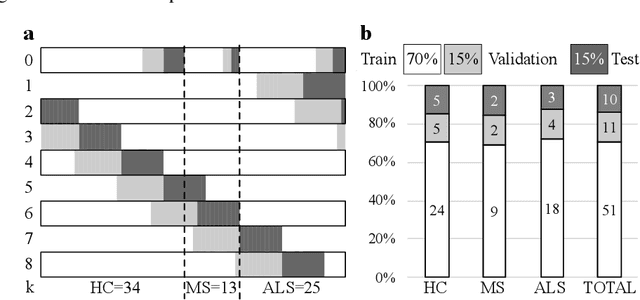
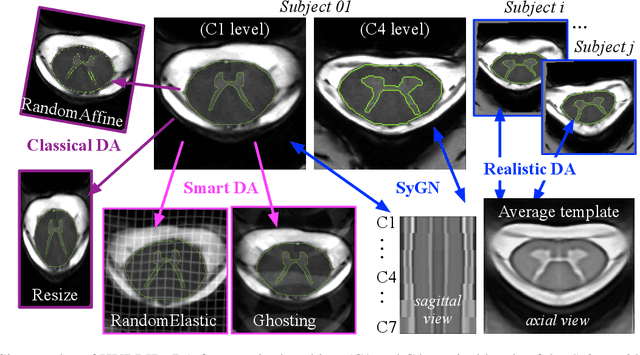
Abstract:The spinal cord (SC), which conveys information between the brain and the peripheral nervous system, plays a key role in various neurological disorders such as multiple sclerosis (MS) and amyotrophic lateral sclerosis (ALS), in which both gray matter (GM) and white matter (WM) may be impaired. While automated methods for WM/GM segmentation are now largely available, these techniques, developed for conventional systems (3T or lower) do not necessarily perform well on 7T MRI data, which feature finer details, contrasts, but also different artifacts or signal dropout. The primary goal of this study is thus to propose a new deep learning model that allows robust SC/GM multi-class segmentation based on ultra-high resolution 7T T2*-w MR images. The second objective is to highlight the relevance of implementing a specific data augmentation (DA) strategy, in particular to generate a generic model that could be used for multi-center studies at 7T.
Team NeuroPoly: Description of the Pipelines for the MICCAI 2021 MS New Lesions Segmentation Challenge
Sep 18, 2021Abstract:This paper gives a detailed description of the pipelines used for the 2nd edition of the MICCAI 2021 Challenge on Multiple Sclerosis Lesion Segmentation. An overview of the data preprocessing steps applied is provided along with a brief description of the pipelines used, in terms of the architecture and the hyperparameters. Our code for this work can be found at: https://github.com/ivadomed/ms-challenge-2021.
Stacked Hourglass Network with a Multi-level Attention Mechanism: Where to Look for Intervertebral Disc Labeling
Aug 14, 2021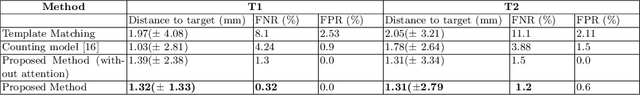
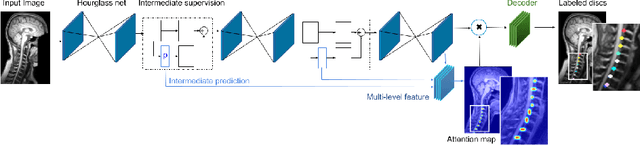
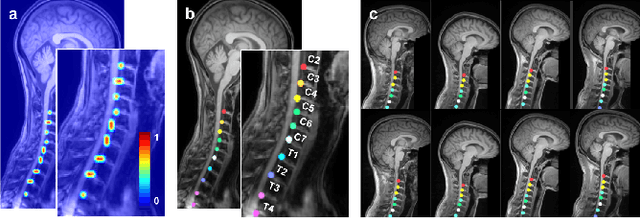
Abstract:Labeling vertebral discs from MRI scans is important for the proper diagnosis of spinal related diseases, including multiple sclerosis, amyotrophic lateral sclerosis, degenerative cervical myelopathy and cancer. Automatic labeling of the vertebral discs in MRI data is a difficult task because of the similarity between discs and bone area, the variability in the geometry of the spine and surrounding tissues across individuals, and the variability across scans (manufacturers, pulse sequence, image contrast, resolution and artefacts). In previous studies, vertebral disc labeling is often done after a disc detection step and mostly fails when the localization algorithm misses discs or has false positive detection. In this work, we aim to mitigate this problem by reformulating the semantic vertebral disc labeling using the pose estimation technique. To do so, we propose a stacked hourglass network with multi-level attention mechanism to jointly learn intervertebral disc position and their skeleton structure. The proposed deep learning model takes into account the strength of semantic segmentation and pose estimation technique to handle the missing area and false positive detection. To further improve the performance of the proposed method, we propose a skeleton-based search space to reduce false positive detection. The proposed method evaluated on spine generic public multi-center dataset and demonstrated better performance comparing to previous work, on both T1w and T2w contrasts. The method is implemented in ivadomed (https://ivadomed.org).
Effectiveness of regional diffusion MRI measures in distinguishing multiple sclerosis abnormalities within the cervical spinal cord
Aug 09, 2021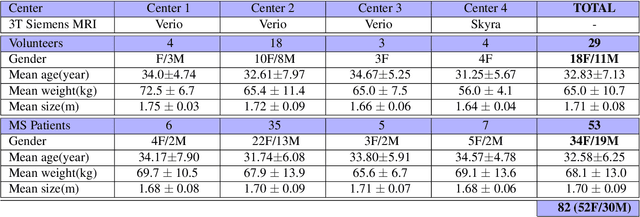
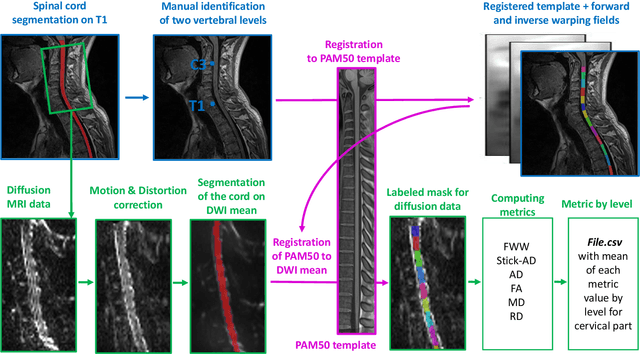
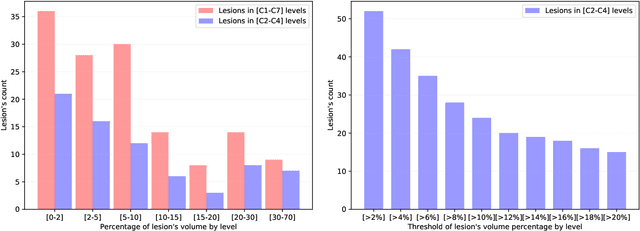
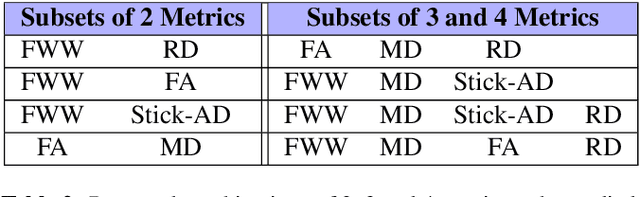
Abstract:Multiple sclerosis is an inflammatory disorder of the central nervous system. Quantitative MRI has huge potential to provide intrinsic and normative values of tissue properties useful for diagnosis, prognosis and ultimately clinical follow-up of this disease. However, there is a large discrepancy between the clinical observations and how the pathology is exhibited in MRI brain scans. Complementary to brain imaging, the study of multiple sclerosis lesions in the spinal cord has recently gained interest as a potential marker for early physical impairment. Therefore, investigating how the spinal cord is damaged using quantitative imaging, in particular, diffusion MRI, becomes an acute challenge. In this work, we extract average diffusion MRI metrics per vertebral level from spinal cord data acquired from multiple clinical sites. The diffusion-based metrics involved are extracted from the diffusion tensor imaging and Ball-and-Stick models and quantified for every cervical vertebral level using a collection of image processing methods and an atlas-based approach. Then, we perform a statistical analysis study to characterize the feasibility of these metrics to detect lesions. Specifically, we study the usefulness of combining different metrics to improve the accuracy prediction score associated with the presence of multiple sclerosis lesions. We demonstrate the grade of sensitivity to underlying microstructure changes in MS patients of each metric. Ball-and-Stick provides novel information about the MS damage to tissue microstructure. In addition, we show that choosing a subset of metrics: [FA, RD, MD] and [FWW, MD, Stick-AD, RD], which bring complementary information, has significantly increased the prediction score of the presence of the MS lesion in the cervical spinal cord.
Evaluation of distortion correction methods in diffusion MRI of the spinal cord
Aug 09, 2021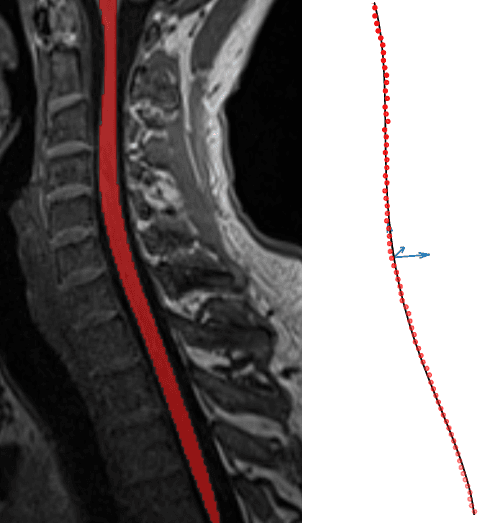
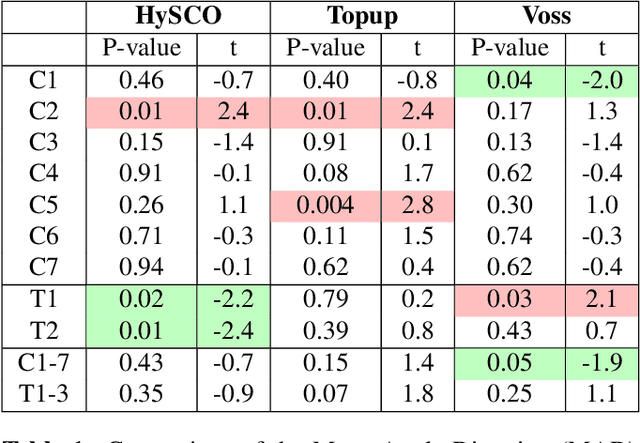
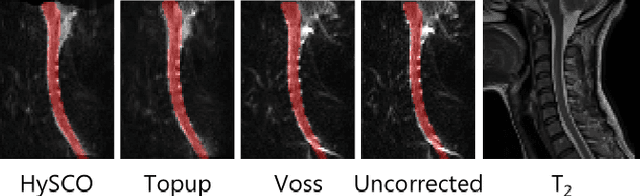
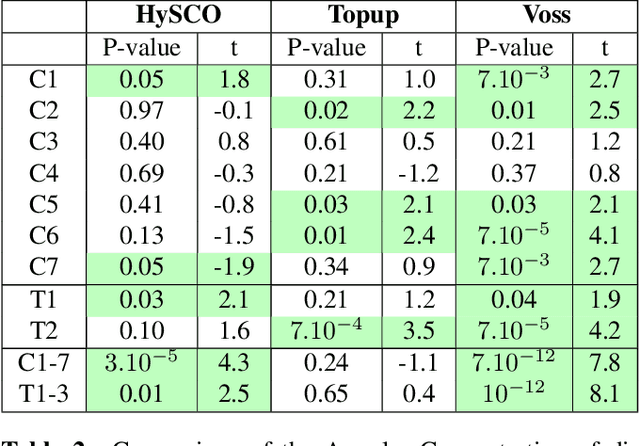
Abstract:Background: Magnetic field inhomogeneities generate important geometric distortions in reconstructed echo-planar images. Various procedures were proposed for correcting these distortions on brain images; yet, few neuroimaging studies tailored and incorporated the use of these techniques in spinal cord diffusion MRI. Purpose: We present a comparative evaluation of distortion correction methods that use the reversed gradient polarity technique on spinal cord. We propose novel geometric metrics to measure the alignment of the reconstructed diffusion model with the apparent centerline of the spinal cord. Subjects: 95 subjects, among which 29 healthy controls and 66 multiple sclerosis patients. Assessment: Geometric distortions were corrected using 4 state-of-the-art methods. We measured the alignment of the principal direction of diffusion with the apparent centerline of the spine after correction and the correlation with the reference anatomical image. Results are computed per vertebral level, to evaluate the impact on different portions of the spine. Besides, subjective evaluation of the quality of the correction of healthy subjects images was performed by three expert raters. Results: As a result of distortion correction, the diffusion directions are better aligned locally with the centerline, in particular at both ends of the acquisition window. The cross-correlation with anatomical image is also improved by Hyperelastic Susceptibility Artefact Correction (HySCO) and block-matching. The subjective evaluation for HySCO is significantly better (p < 0.05) than for Block-Matching; TOPUP performs significantly worse than the three other methods. Conclusion: Correction based on HySCO provide best results among the selected methods.
Reproducibility and Evolution of Diffusion MRI Measurements within the Cervical Spinal Cord in Multiple Sclerosis
Aug 08, 2021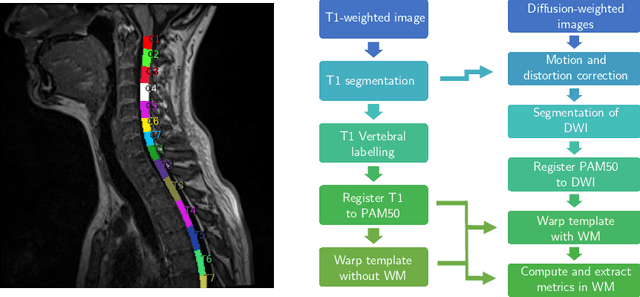
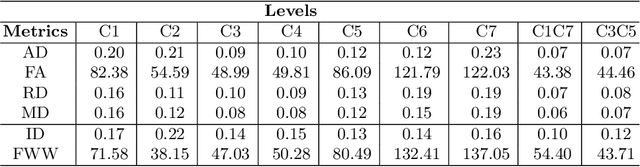
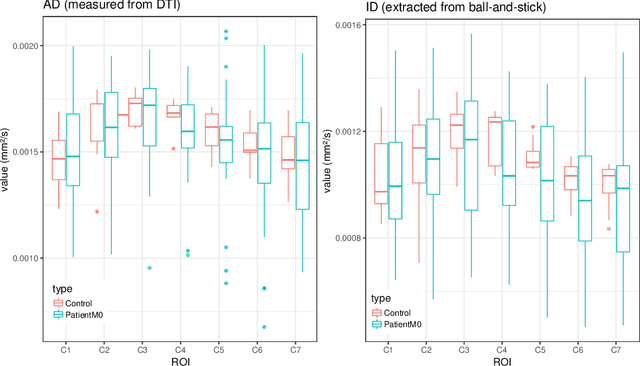
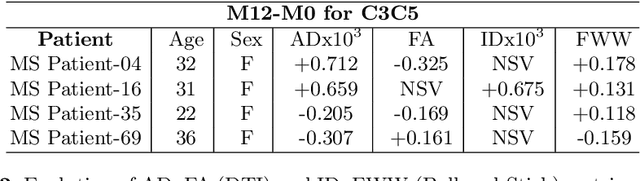
Abstract:In Multiple Sclerosis (MS), there is a large discrepancy between the clinical observations and how the pathology is exhibited on brain images, this is known as the clinical-radiological paradox (CRP). One of the hypotheses is that the clinical deficit may be more related to the spinal cord damage than the number or location of lesions in the brain. Therefore, investigating how the spinal cord is damaged becomes an acute challenge to better understand and overcome the CRP. Diffusion MRI is known to provide quantitative figures of neuronal degeneration and axonal loss, in the brain as well as in the spinal cord. In this paper, we propose to investigate how diffusion MRI metrics vary in the different cervical regions with the progression of the disease. We first study the reproducibility of diffusion MRI on healthy volunteers with a test-retest procedure using both standard diffusion tensor imaging (DTI) and multi-compartment Ball-and-Stick models. Then, based on the test re-test quantitative calibration, we provide quantitative figures of pathology evolution between M0 and M12 in the cervical spine on a set of 31 MS patients, exhibiting how the pathology damage spans in the cervical spinal cord.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge