Charley Gros
Label fusion and training methods for reliable representation of inter-rater uncertainty
Feb 26, 2022
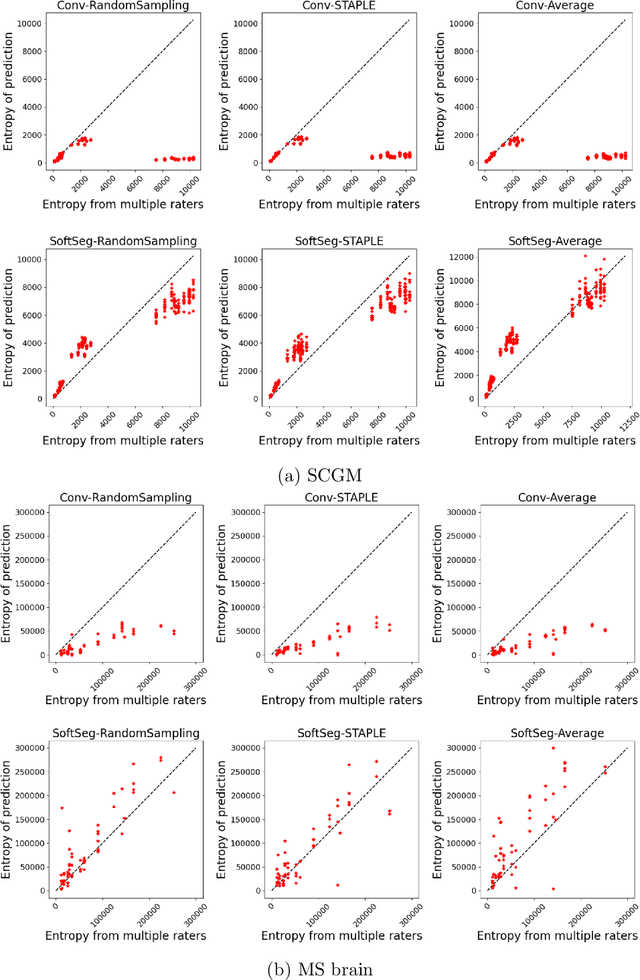
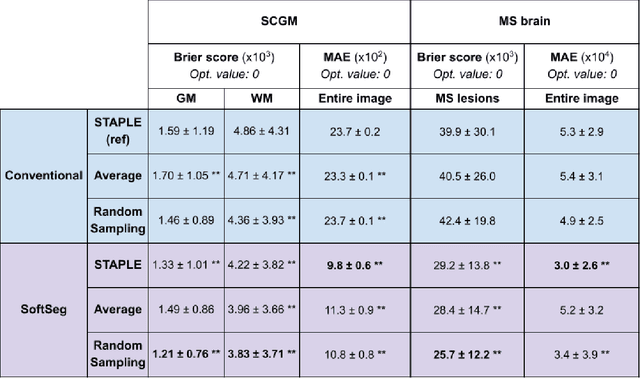
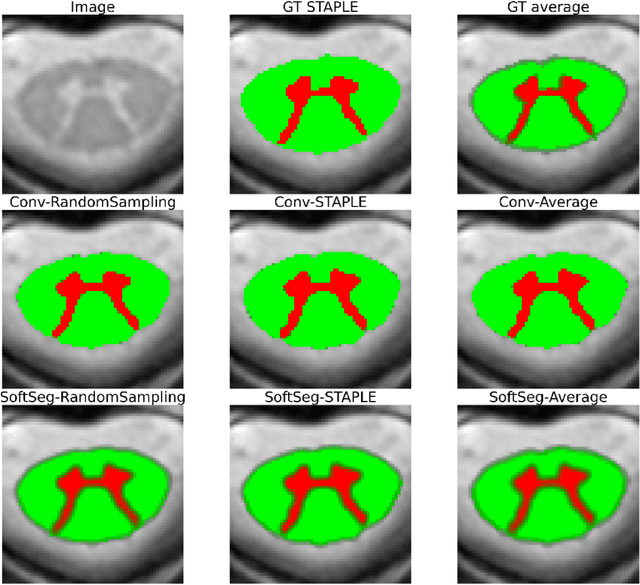
Abstract:Medical tasks are prone to inter-rater variability due to multiple factors such as image quality, professional experience and training, or guideline clarity. Training deep learning networks with annotations from multiple raters is a common practice that mitigates the model's bias towards a single expert. Reliable models generating calibrated outputs and reflecting the inter-rater disagreement are key to the integration of artificial intelligence in clinical practice. Various methods exist to take into account different expert labels. We focus on comparing three label fusion methods: STAPLE, average of the rater's segmentation, and random sampling of each rater's segmentation during training. Each label fusion method is studied using both the conventional training framework and the recently published SoftSeg framework that limits information loss by treating the segmentation task as a regression. Our results, across 10 data splittings on two public datasets, indicate that SoftSeg models, regardless of the ground truth fusion method, had better calibration and preservation of the inter-rater rater variability compared with their conventional counterparts without impacting the segmentation performance. Conventional models, i.e., trained with a Dice loss, with binary inputs, and sigmoid/softmax final activate, were overconfident and underestimated the uncertainty associated with inter-rater variability. Conversely, fusing labels by averaging with the SoftSeg framework led to underconfident outputs and overestimation of the rater disagreement. In terms of segmentation performance, the best label fusion method was different for the two datasets studied, indicating this parameter might be task-dependent. However, SoftSeg had segmentation performance systematically superior or equal to the conventionally trained models and had the best calibration and preservation of the inter-rater variability.
2D Multi-Class Model for Gray and White Matter Segmentation of the Cervical Spinal Cord at 7T
Oct 13, 2021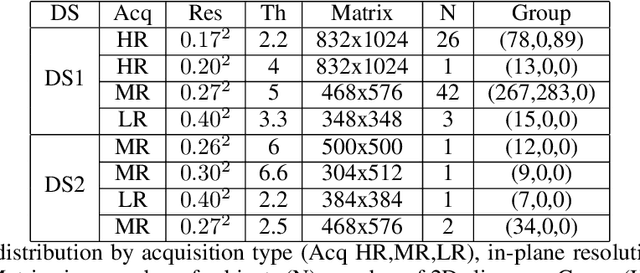
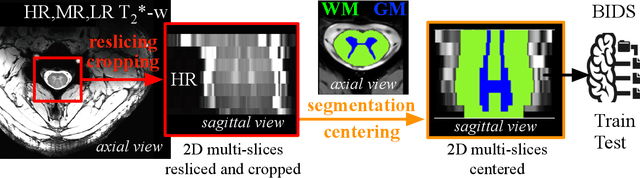
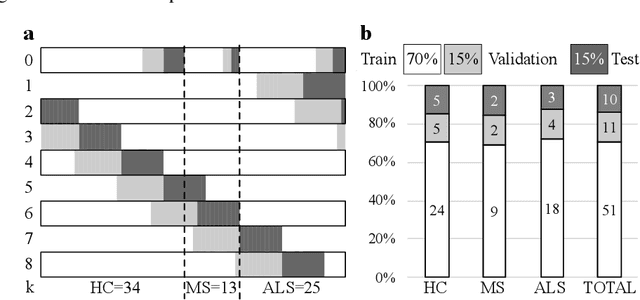
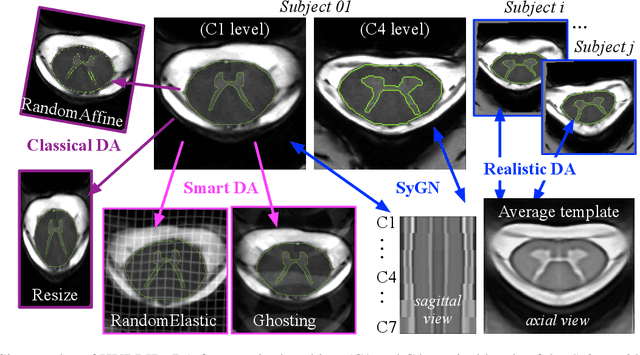
Abstract:The spinal cord (SC), which conveys information between the brain and the peripheral nervous system, plays a key role in various neurological disorders such as multiple sclerosis (MS) and amyotrophic lateral sclerosis (ALS), in which both gray matter (GM) and white matter (WM) may be impaired. While automated methods for WM/GM segmentation are now largely available, these techniques, developed for conventional systems (3T or lower) do not necessarily perform well on 7T MRI data, which feature finer details, contrasts, but also different artifacts or signal dropout. The primary goal of this study is thus to propose a new deep learning model that allows robust SC/GM multi-class segmentation based on ultra-high resolution 7T T2*-w MR images. The second objective is to highlight the relevance of implementing a specific data augmentation (DA) strategy, in particular to generate a generic model that could be used for multi-center studies at 7T.
Team NeuroPoly: Description of the Pipelines for the MICCAI 2021 MS New Lesions Segmentation Challenge
Sep 18, 2021Abstract:This paper gives a detailed description of the pipelines used for the 2nd edition of the MICCAI 2021 Challenge on Multiple Sclerosis Lesion Segmentation. An overview of the data preprocessing steps applied is provided along with a brief description of the pipelines used, in terms of the architecture and the hyperparameters. Our code for this work can be found at: https://github.com/ivadomed/ms-challenge-2021.
Impact of individual rater style on deep learning uncertainty in medical imaging segmentation
May 05, 2021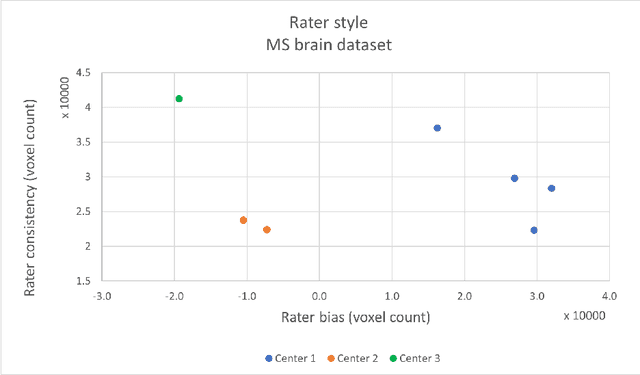
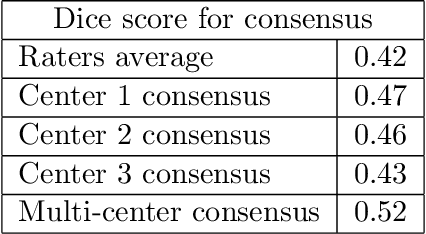
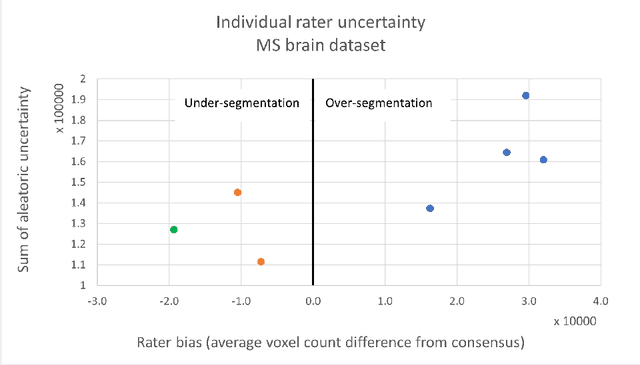
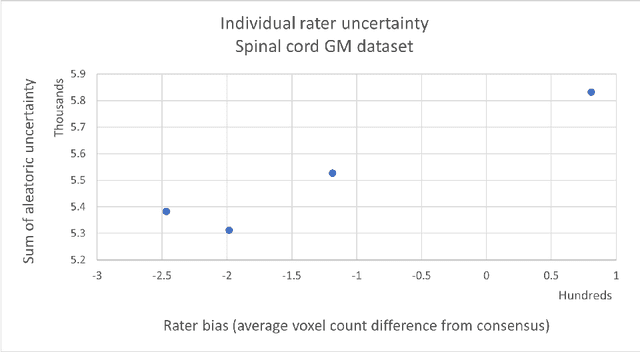
Abstract:While multiple studies have explored the relation between inter-rater variability and deep learning model uncertainty in medical segmentation tasks, little is known about the impact of individual rater style. This study quantifies rater style in the form of bias and consistency and explores their impacts when used to train deep learning models. Two multi-rater public datasets were used, consisting of brain multiple sclerosis lesion and spinal cord grey matter segmentation. On both datasets, results show a correlation ($R^2 = 0.60$ and $0.93$) between rater bias and deep learning uncertainty. The impact of label fusion between raters' annotations on this relationship is also explored, and we show that multi-center consensuses are more effective than single-center consensuses to reduce uncertainty, since rater style is mostly center-specific.
Benefits of Linear Conditioning for Segmentation using Metadata
Feb 18, 2021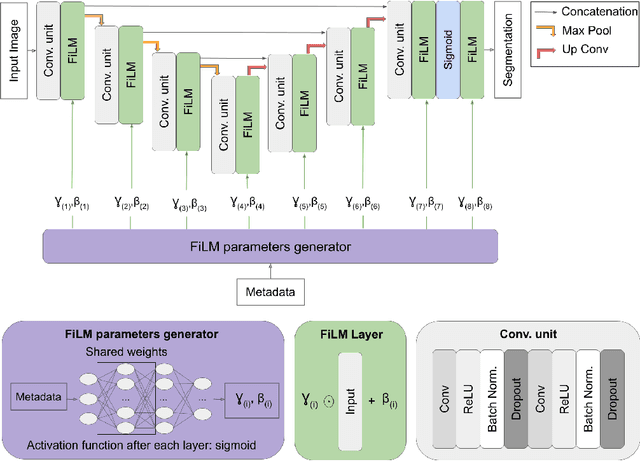
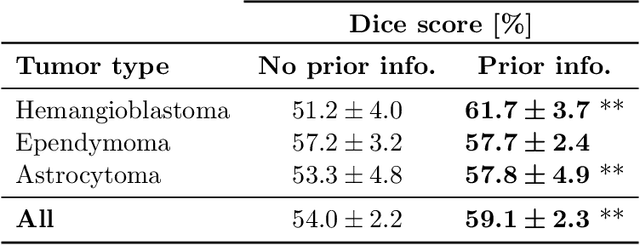
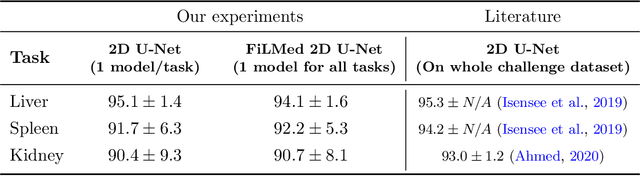
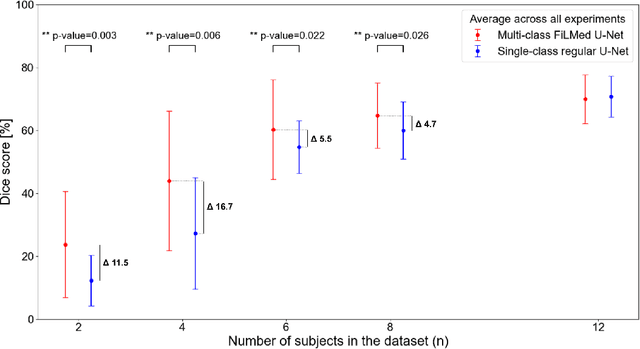
Abstract:Medical images are often accompanied by metadata describing the image (vendor, acquisition parameters) and the patient (disease type or severity, demographics, genomics). This metadata is usually disregarded by image segmentation methods. In this work, we adapt a linear conditioning method called FiLM (Feature-wise Linear Modulation) for image segmentation tasks. This FiLM adaptation enables integrating metadata into segmentation models for better performance. We observed an average Dice score increase of 5.1% on spinal cord tumor segmentation when incorporating the tumor type with FiLM. The metadata modulates the segmentation process through low-cost affine transformations applied on feature maps which can be included in any neural network's architecture. Additionally, we assess the relevance of segmentation FiLM layers for tackling common challenges in medical imaging: training with limited or unbalanced number of annotated data, multi-class training with missing segmentations, and model adaptation to multiple tasks. Our results demonstrated the following benefits of FiLM for segmentation: FiLMed U-Net was robust to missing labels and reached higher Dice scores with few labels (up to 16.7%) compared to single-task U-Net. The code is open-source and available at www.ivadomed.org.
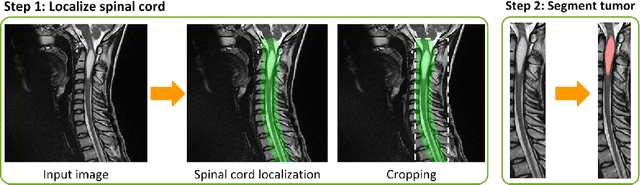
Multiclass Spinal Cord Tumor Segmentation on MRI with Deep Learning
Jan 14, 2021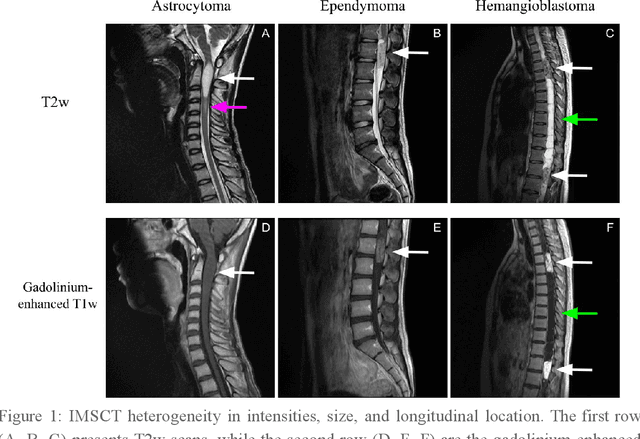
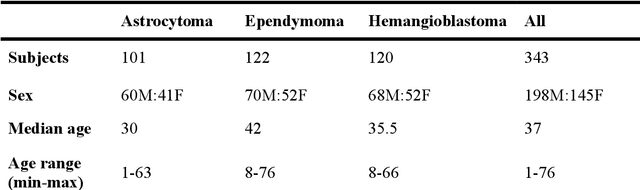
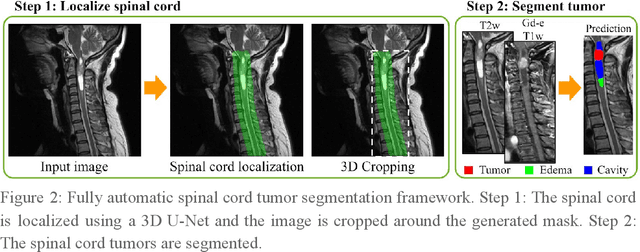
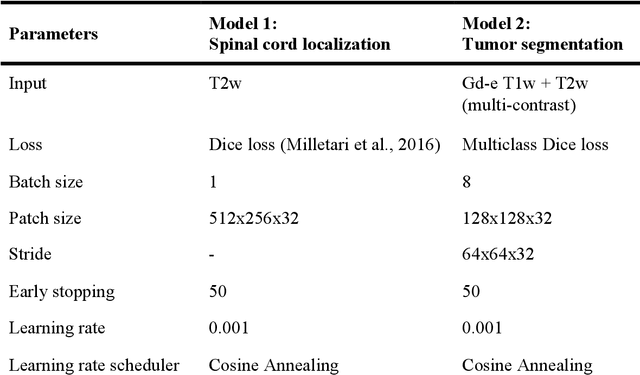
Abstract:Spinal cord tumors lead to neurological morbidity and mortality. Being able to obtain morphometric quantification (size, location, growth rate) of the tumor, edema, and cavity can result in improved monitoring and treatment planning. Such quantification requires the segmentation of these structures into three separate classes. However, manual segmentation of 3-dimensional structures is time-consuming and tedious, motivating the development of automated methods. Here, we tailor a model adapted to the spinal cord tumor segmentation task. Data were obtained from 343 patients using gadolinium-enhanced T1-weighted and T2-weighted MRI scans with cervical, thoracic, and/or lumbar coverage. The dataset includes the three most common intramedullary spinal cord tumor types: astrocytomas, ependymomas, and hemangioblastomas. The proposed approach is a cascaded architecture with U-Net-based models that segments tumors in a two-stage process: locate and label. The model first finds the spinal cord and generates bounding box coordinates. The images are cropped according to this output, leading to a reduced field of view, which mitigates class imbalance. The tumor is then segmented. The segmentation of the tumor, cavity, and edema (as a single class) reached 76.7 $\pm$ 1.5% of Dice score and the segmentation of tumors alone reached 61.8 $\pm$ 4.0% Dice score. The true positive detection rate was above 87% for tumor, edema, and cavity. To the best of our knowledge, this is the first fully automatic deep learning model for spinal cord tumor segmentation. The multiclass segmentation pipeline is available in the Spinal Cord Toolbox (https://spinalcordtoolbox.com/). It can be run with custom data on a regular computer within seconds.
SoftSeg: Advantages of soft versus binary training for image segmentation
Nov 18, 2020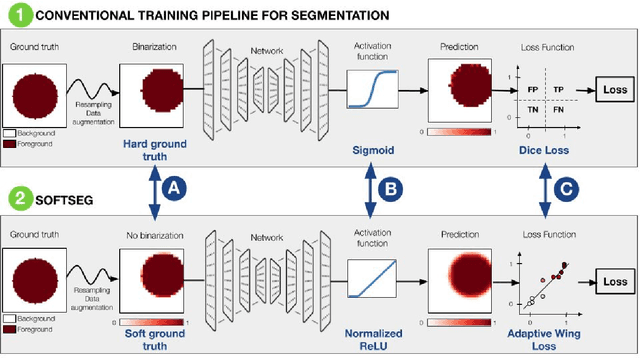
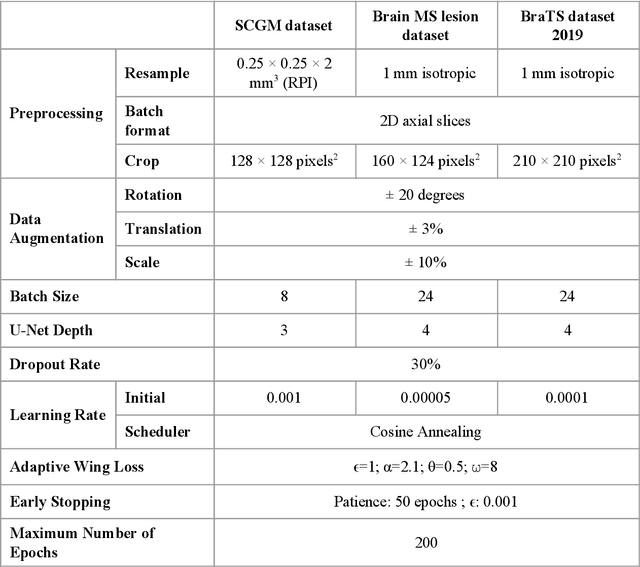
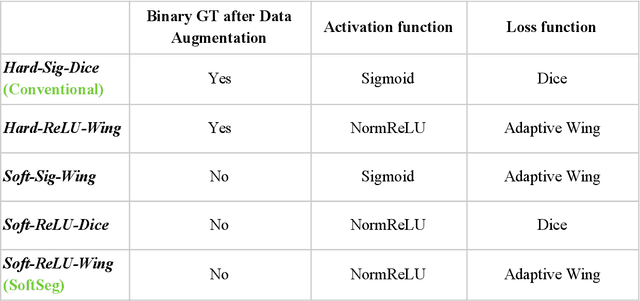
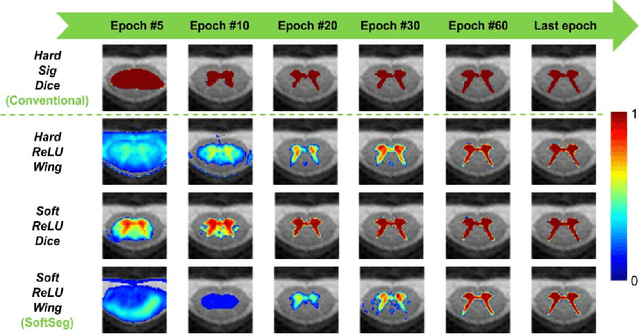
Abstract:Most image segmentation algorithms are trained on binary masks formulated as a classification task per pixel. However, in applications such as medical imaging, this "black-and-white" approach is too constraining because the contrast between two tissues is often ill-defined, i.e., the voxels located on objects' edges contain a mixture of tissues. Consequently, assigning a single "hard" label can result in a detrimental approximation. Instead, a soft prediction containing non-binary values would overcome that limitation. We introduce SoftSeg, a deep learning training approach that takes advantage of soft ground truth labels, and is not bound to binary predictions. SoftSeg aims at solving a regression instead of a classification problem. This is achieved by using (i) no binarization after preprocessing and data augmentation, (ii) a normalized ReLU final activation layer (instead of sigmoid), and (iii) a regression loss function (instead of the traditional Dice loss). We assess the impact of these three features on three open-source MRI segmentation datasets from the spinal cord gray matter, the multiple sclerosis brain lesion, and the multimodal brain tumor segmentation challenges. Across multiple cross-validation iterations, SoftSeg outperformed the conventional approach, leading to an increase in Dice score of 2.0% on the gray matter dataset (p=0.001), 3.3% for the MS lesions, and 6.5% for the brain tumors. SoftSeg produces consistent soft predictions at tissues' interfaces and shows an increased sensitivity for small objects. The richness of soft labels could represent the inter-expert variability, the partial volume effect, and complement the model uncertainty estimation. The developed training pipeline can easily be incorporated into most of the existing deep learning architectures. It is already implemented in the freely-available deep learning toolbox ivadomed (https://ivadomed.org).
ivadomed: A Medical Imaging Deep Learning Toolbox
Oct 20, 2020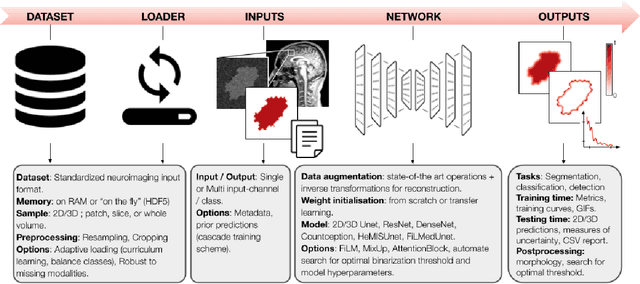
Abstract:ivadomed is an open-source Python package for designing, end-to-end training, and evaluating deep learning models applied to medical imaging data. The package includes APIs, command-line tools, documentation, and tutorials. ivadomed also includes pre-trained models such as spinal tumor segmentation and vertebral labeling. Original features of ivadomed include a data loader that can parse image metadata (e.g., acquisition parameters, image contrast, resolution) and subject metadata (e.g., pathology, age, sex) for custom data splitting or extra information during training and evaluation. Any dataset following the Brain Imaging Data Structure (BIDS) convention will be compatible with ivadomed without the need to manually organize the data, which is typically a tedious task. Beyond the traditional deep learning methods, ivadomed features cutting-edge architectures, such as FiLM and HeMis, as well as various uncertainty estimation methods (aleatoric and epistemic), and losses adapted to imbalanced classes and non-binary predictions. Each step is conveniently configurable via a single file. At the same time, the code is highly modular to allow addition/modification of an architecture or pre/post-processing steps. Example applications of ivadomed include MRI object detection, segmentation, and labeling of anatomical and pathological structures. Overall, ivadomed enables easy and quick exploration of the latest advances in deep learning for medical imaging applications. ivadomed's main project page is available at https://ivadomed.org.
Automatic segmentation of spinal multiple sclerosis lesions: How to generalize across MRI contrasts?
Mar 11, 2020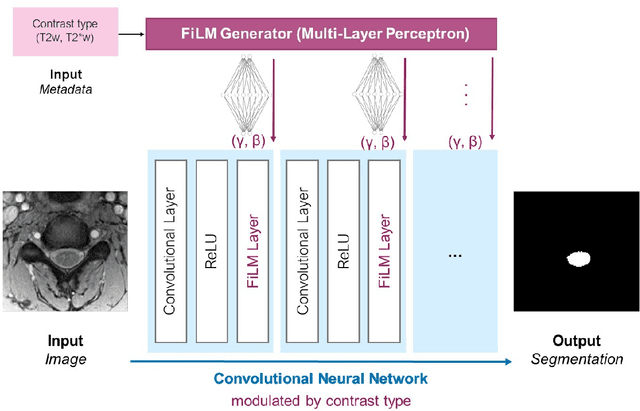
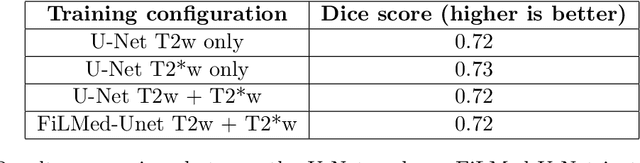
Abstract:Despite recent improvements in medical image segmentation, the ability to generalize across imaging contrasts remains an open issue. To tackle this challenge, we implement Feature-wise Linear Modulation (FiLM) to leverage physics knowledge within the segmentation model and learn the characteristics of each contrast. Interestingly, a well-optimised U-Net reached the same performance as our FiLMed-Unet on a multi-contrast dataset (0.72 of Dice score), which suggests that there is a bottleneck in spinal MS lesion segmentation different from the generalization across varying contrasts. This bottleneck likely stems from inter-rater variability, which is estimated at 0.61 of Dice score in our dataset.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018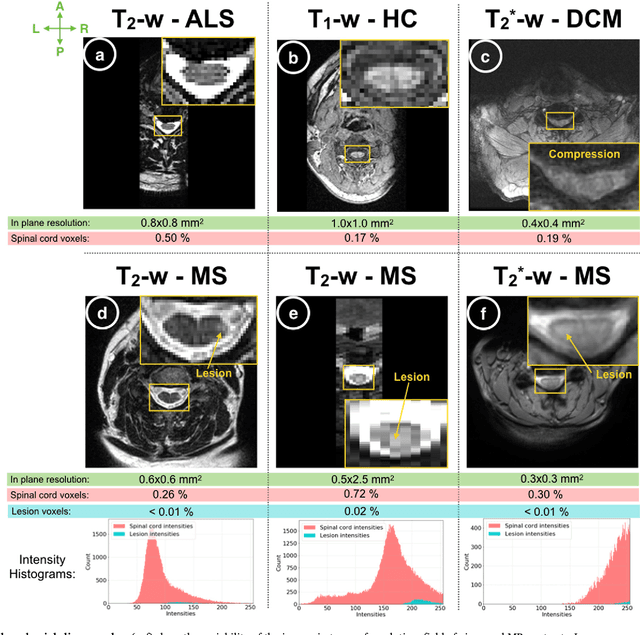
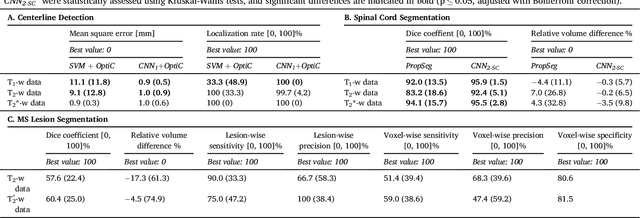
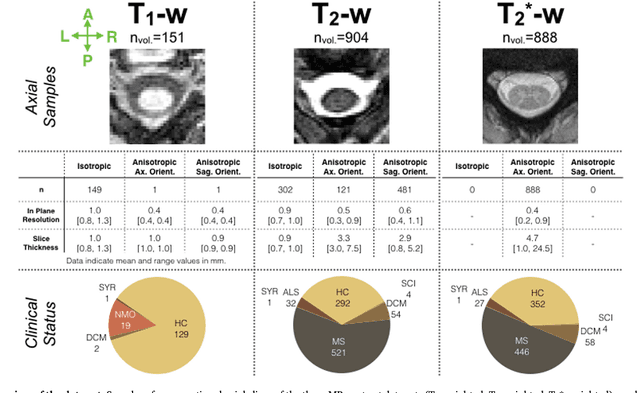
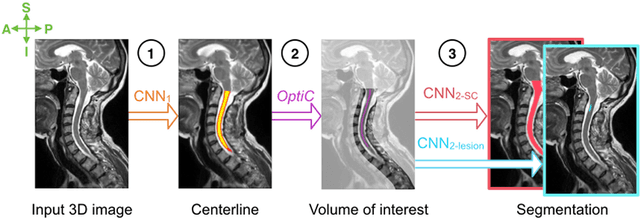
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge