Nika Khosravi
SMU-Net: Style matching U-Net for brain tumor segmentation with missing modalities
Apr 06, 2022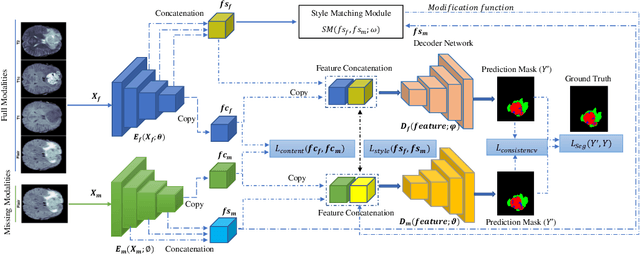
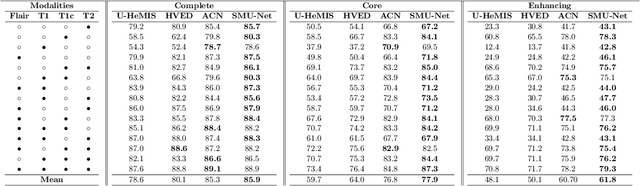
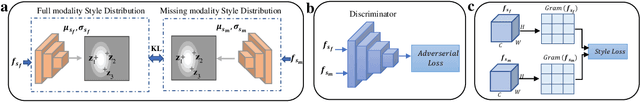
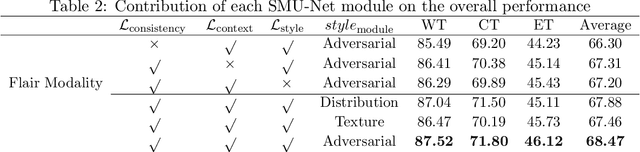
Abstract:Gliomas are one of the most prevalent types of primary brain tumours, accounting for more than 30\% of all cases and they develop from the glial stem or progenitor cells. In theory, the majority of brain tumours could well be identified exclusively by the use of Magnetic Resonance Imaging (MRI). Each MRI modality delivers distinct information on the soft tissue of the human brain and integrating all of them would provide comprehensive data for the accurate segmentation of the glioma, which is crucial for the patient's prognosis, diagnosis, and determining the best follow-up treatment. Unfortunately, MRI is prone to artifacts for a variety of reasons, which might result in missing one or more MRI modalities. Various strategies have been proposed over the years to synthesize the missing modality or compensate for the influence it has on automated segmentation models. However, these methods usually fail to model the underlying missing information. In this paper, we propose a style matching U-Net (SMU-Net) for brain tumour segmentation on MRI images. Our co-training approach utilizes a content and style-matching mechanism to distill the informative features from the full-modality network into a missing modality network. To do so, we encode both full-modality and missing-modality data into a latent space, then we decompose the representation space into a style and content representation. Our style matching module adaptively recalibrates the representation space by learning a matching function to transfer the informative and textural features from a full-modality path into a missing-modality path. Moreover, by modelling the mutual information, our content module surpasses the less informative features and re-calibrates the representation space based on discriminative semantic features. The evaluation process on the BraTS 2018 dataset shows a significant results.
Medical Image Segmentation on MRI Images with Missing Modalities: A Review
Mar 11, 2022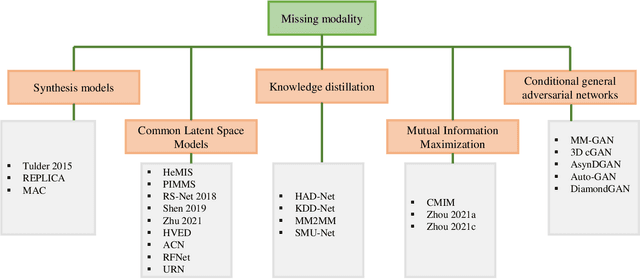
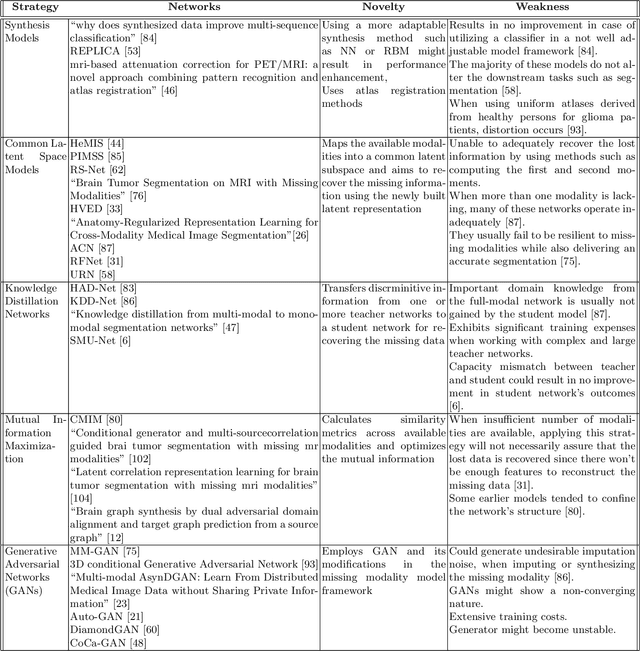
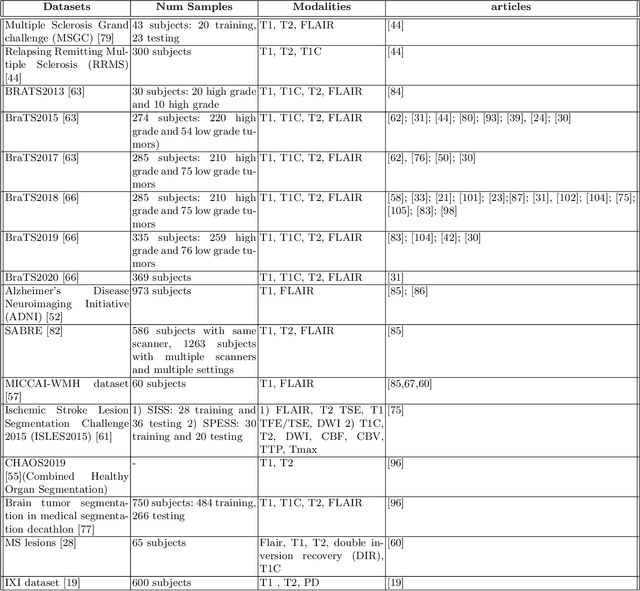
Abstract:Dealing with missing modalities in Magnetic Resonance Imaging (MRI) and overcoming their negative repercussions is considered a hurdle in biomedical imaging. The combination of a specified set of modalities, which is selected depending on the scenario and anatomical part being scanned, will provide medical practitioners with full information about the region of interest in the human body, hence the missing MRI sequences should be reimbursed. The compensation of the adverse impact of losing useful information owing to the lack of one or more modalities is a well-known challenge in the field of computer vision, particularly for medical image processing tasks including tumour segmentation, tissue classification, and image generation. Various approaches have been developed over time to mitigate this problem's negative implications and this literature review goes through a significant number of the networks that seek to do so. The approaches reviewed in this work are reviewed in detail, including earlier techniques such as synthesis methods as well as later approaches that deploy deep learning, such as common latent space models, knowledge distillation networks, mutual information maximization, and generative adversarial networks (GANs). This work discusses the most important approaches that have been offered at the time of this writing, examining the novelty, strength, and weakness of each one. Furthermore, the most commonly used MRI datasets are highlighted and described. The main goal of this research is to offer a performance evaluation of missing modality compensating networks, as well as to outline future strategies for dealing with this issue.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge