Lucas Rouhier
Stacked Hourglass Network with a Multi-level Attention Mechanism: Where to Look for Intervertebral Disc Labeling
Aug 14, 2021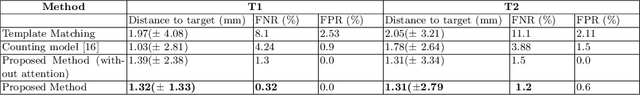
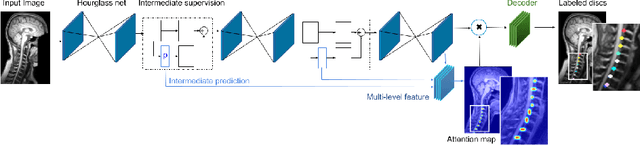
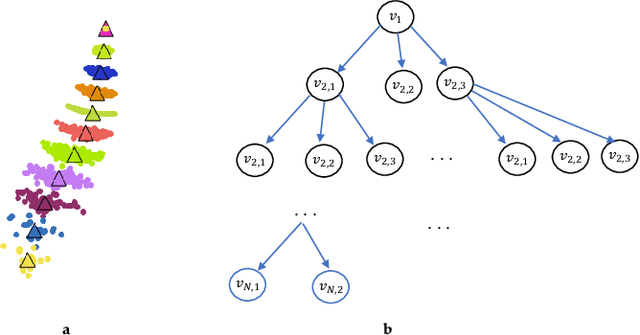
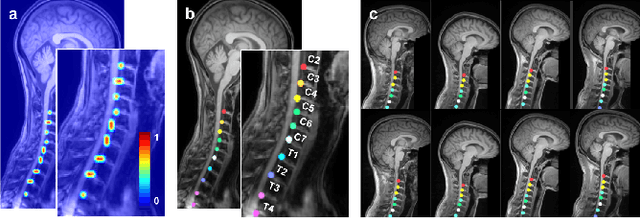
Abstract:Labeling vertebral discs from MRI scans is important for the proper diagnosis of spinal related diseases, including multiple sclerosis, amyotrophic lateral sclerosis, degenerative cervical myelopathy and cancer. Automatic labeling of the vertebral discs in MRI data is a difficult task because of the similarity between discs and bone area, the variability in the geometry of the spine and surrounding tissues across individuals, and the variability across scans (manufacturers, pulse sequence, image contrast, resolution and artefacts). In previous studies, vertebral disc labeling is often done after a disc detection step and mostly fails when the localization algorithm misses discs or has false positive detection. In this work, we aim to mitigate this problem by reformulating the semantic vertebral disc labeling using the pose estimation technique. To do so, we propose a stacked hourglass network with multi-level attention mechanism to jointly learn intervertebral disc position and their skeleton structure. The proposed deep learning model takes into account the strength of semantic segmentation and pose estimation technique to handle the missing area and false positive detection. To further improve the performance of the proposed method, we propose a skeleton-based search space to reduce false positive detection. The proposed method evaluated on spine generic public multi-center dataset and demonstrated better performance comparing to previous work, on both T1w and T2w contrasts. The method is implemented in ivadomed (https://ivadomed.org).
ivadomed: A Medical Imaging Deep Learning Toolbox
Oct 20, 2020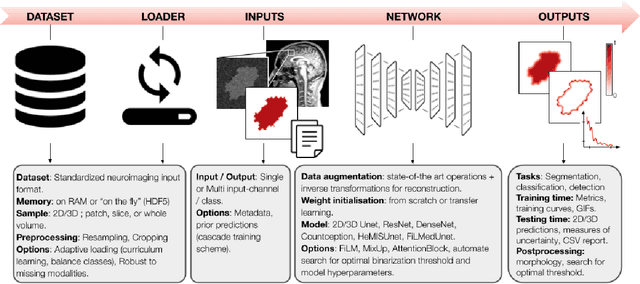
Abstract:ivadomed is an open-source Python package for designing, end-to-end training, and evaluating deep learning models applied to medical imaging data. The package includes APIs, command-line tools, documentation, and tutorials. ivadomed also includes pre-trained models such as spinal tumor segmentation and vertebral labeling. Original features of ivadomed include a data loader that can parse image metadata (e.g., acquisition parameters, image contrast, resolution) and subject metadata (e.g., pathology, age, sex) for custom data splitting or extra information during training and evaluation. Any dataset following the Brain Imaging Data Structure (BIDS) convention will be compatible with ivadomed without the need to manually organize the data, which is typically a tedious task. Beyond the traditional deep learning methods, ivadomed features cutting-edge architectures, such as FiLM and HeMis, as well as various uncertainty estimation methods (aleatoric and epistemic), and losses adapted to imbalanced classes and non-binary predictions. Each step is conveniently configurable via a single file. At the same time, the code is highly modular to allow addition/modification of an architecture or pre/post-processing steps. Example applications of ivadomed include MRI object detection, segmentation, and labeling of anatomical and pathological structures. Overall, ivadomed enables easy and quick exploration of the latest advances in deep learning for medical imaging applications. ivadomed's main project page is available at https://ivadomed.org.
Spine intervertebral disc labeling using a fully convolutional redundant counting model
Mar 11, 2020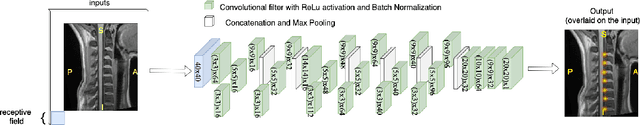
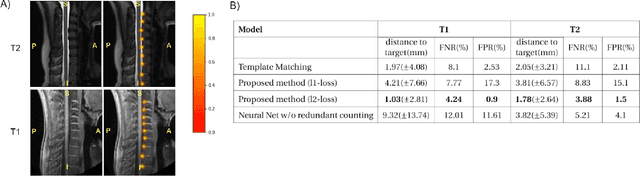
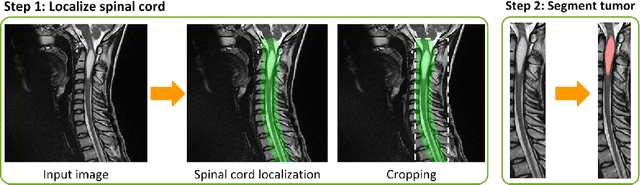
Abstract:Labeling intervertebral discs is relevant as it notably enables clinicians to understand the relationship between a patient's symptoms (pain, paralysis) and the exact level of spinal cord injury. However manually labeling those discs is a tedious and user-biased task which would benefit from automated methods. While some automated methods already exist for MRI and CT-scan, they are either not publicly available, or fail to generalize across various imaging contrasts. In this paper we combine a Fully Convolutional Network (FCN) with inception modules to localize and label intervertebral discs. We demonstrate a proof-of-concept application in a publicly-available multi-center and multi-contrast MRI database (n=235 subjects). The code is publicly available at https://github.com/neuropoly/vertebral-labeling-deep-learning.
U-Net Fixed-Point Quantization for Medical Image Segmentation
Sep 09, 2019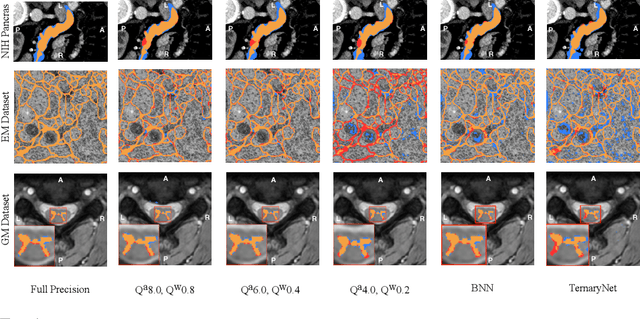
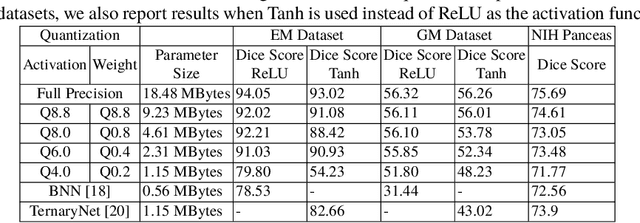
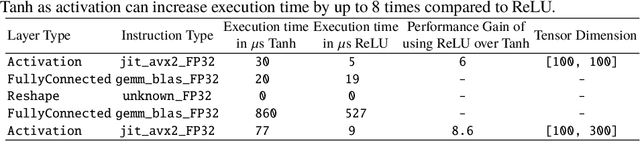
Abstract:Model quantization is leveraged to reduce the memory consumption and the computation time of deep neural networks. This is achieved by representing weights and activations with a lower bit resolution when compared to their high precision floating point counterparts. The suitable level of quantization is directly related to the model performance. Lowering the quantization precision (e.g. 2 bits), reduces the amount of memory required to store model parameters and the amount of logic required to implement computational blocks, which contributes to reducing the power consumption of the entire system. These benefits typically come at the cost of reduced accuracy. The main challenge is to quantize a network as much as possible, while maintaining the performance accuracy. In this work, we present a quantization method for the U-Net architecture, a popular model in medical image segmentation. We then apply our quantization algorithm to three datasets: (1) the Spinal Cord Gray Matter Segmentation (GM), (2) the ISBI challenge for segmentation of neuronal structures in Electron Microscopic (EM), and (3) the public National Institute of Health (NIH) dataset for pancreas segmentation in abdominal CT scans. The reported results demonstrate that with only 4 bits for weights and 6 bits for activations, we obtain 8 fold reduction in memory requirements while loosing only 2.21%, 0.57% and 2.09% dice overlap score for EM, GM and NIH datasets respectively. Our fixed point quantization provides a flexible trade off between accuracy and memory requirement which is not provided by previous quantization methods for U-Net such as TernaryNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge