José Miguel Hernández-Lobato
Bayesian Experimental Design for Computed Tomography with the Linearised Deep Image Prior
Jul 11, 2022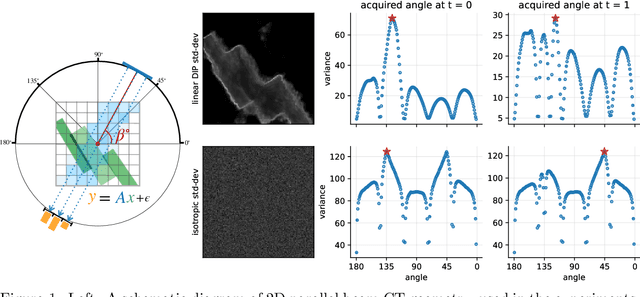
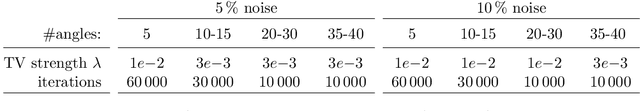
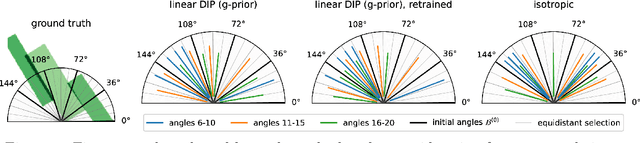

Abstract:We investigate adaptive design based on a single sparse pilot scan for generating effective scanning strategies for computed tomography reconstruction. We propose a novel approach using the linearised deep image prior. It allows incorporating information from the pilot measurements into the angle selection criteria, while maintaining the tractability of a conjugate Gaussian-linear model. On a synthetically generated dataset with preferential directions, linearised DIP design allows reducing the number of scans by up to 30% relative to an equidistant angle baseline.
Adapting the Linearised Laplace Model Evidence for Modern Deep Learning
Jun 17, 2022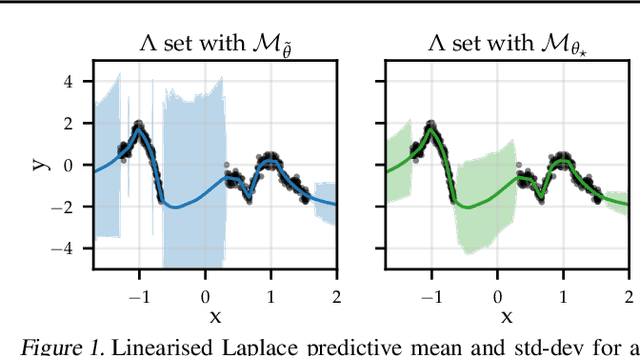
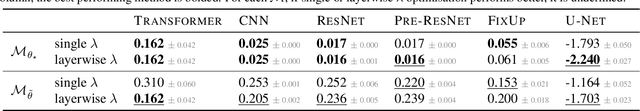
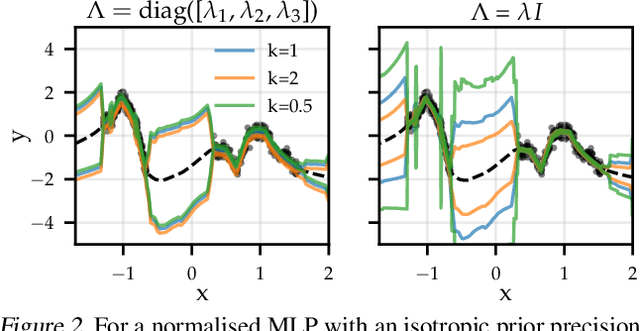
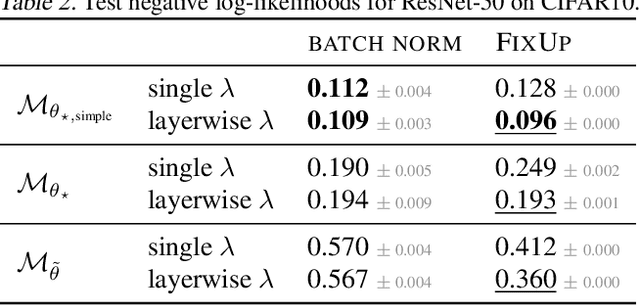
Abstract:The linearised Laplace method for estimating model uncertainty has received renewed attention in the Bayesian deep learning community. The method provides reliable error bars and admits a closed-form expression for the model evidence, allowing for scalable selection of model hyperparameters. In this work, we examine the assumptions behind this method, particularly in conjunction with model selection. We show that these interact poorly with some now-standard tools of deep learning--stochastic approximation methods and normalisation layers--and make recommendations for how to better adapt this classic method to the modern setting. We provide theoretical support for our recommendations and validate them empirically on MLPs, classic CNNs, residual networks with and without normalisation layers, generative autoencoders and transformers.
Meta-learning Feature Representations for Adaptive Gaussian Processes via Implicit Differentiation
May 05, 2022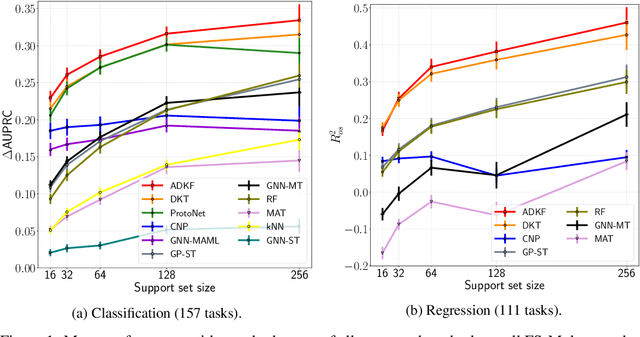
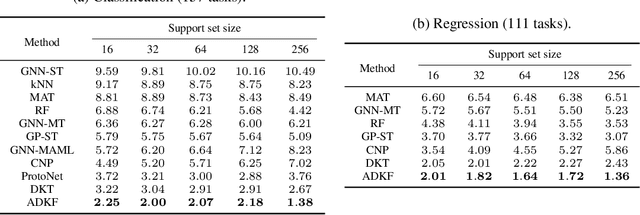
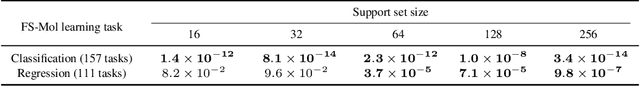
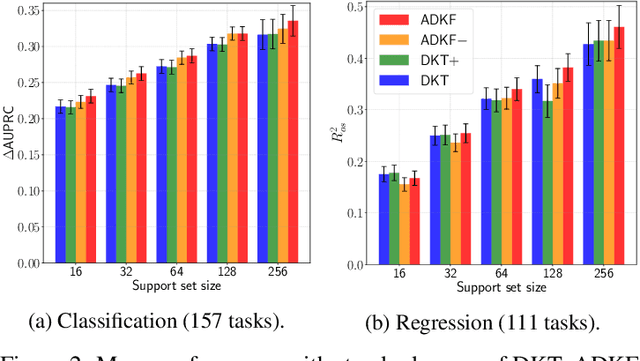
Abstract:We propose Adaptive Deep Kernel Fitting (ADKF), a general framework for learning deep kernels by interpolating between meta-learning and conventional learning. Our approach employs a bilevel optimization objective where we meta-learn feature representations that are generally useful across tasks, in the sense that task-specific Gaussian process models estimated on top of such features achieve the lowest possible predictive loss on average across tasks. We solve the resulting nested optimization problem using the implicit function theorem. We show that ADKF contains Deep Kernel Learning and Deep Kernel Transfer as special cases. Although ADKF is a completely general method, we argue that it is especially well-suited for drug discovery problems and demonstrate that it significantly outperforms previous state-of-the-art methods on a variety of real-world few-shot molecular property prediction tasks and out-of-domain molecular optimization tasks.
A Probabilistic Deep Image Prior for Computational Tomography
Feb 28, 2022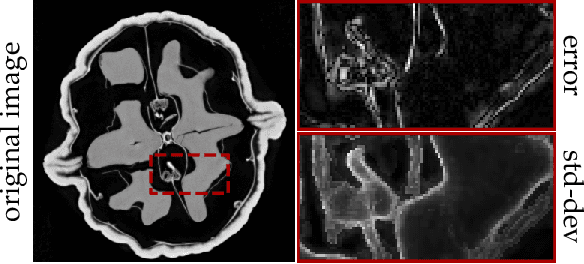
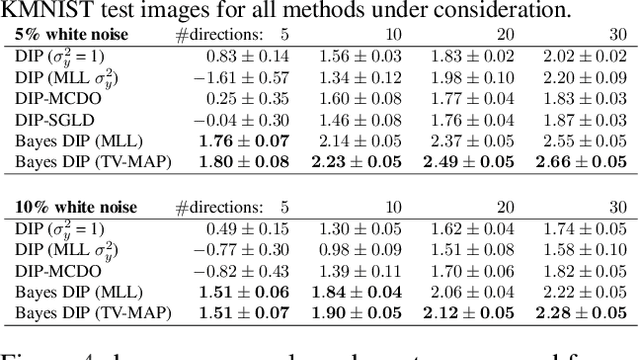
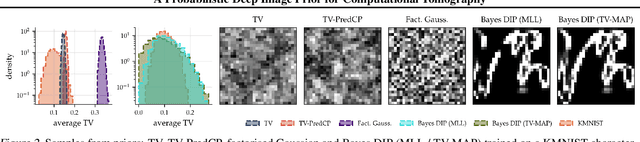
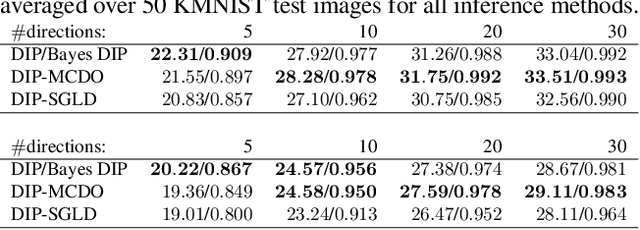
Abstract:Existing deep-learning based tomographic image reconstruction methods do not provide accurate estimates of reconstruction uncertainty, hindering their real-world deployment. To address this limitation, we construct a Bayesian prior for tomographic reconstruction, which combines the classical total variation (TV) regulariser with the modern deep image prior (DIP). Specifically, we use a change of variables to connect our prior beliefs on the image TV semi-norm with the hyper-parameters of the DIP network. For the inference, we develop an approach based on the linearised Laplace method, which is scalable to high-dimensional settings. The resulting framework provides pixel-wise uncertainty estimates and a marginal likelihood objective for hyperparameter optimisation. We demonstrate the method on synthetic and real-measured high-resolution $\mu$CT data, and show that it provides superior calibration of uncertainty estimates relative to previous probabilistic formulations of the DIP.
Missing Data Imputation and Acquisition with Deep Hierarchical Models and Hamiltonian Monte Carlo
Feb 09, 2022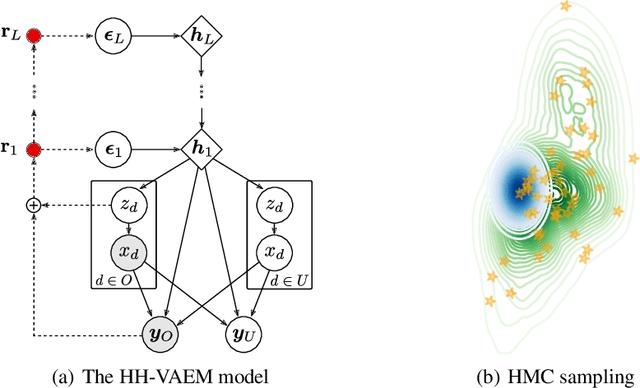


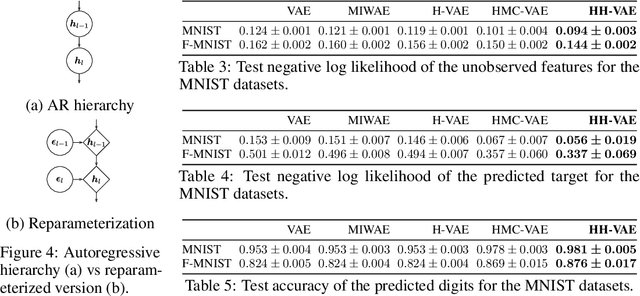
Abstract:Variational Autoencoders (VAEs) have recently been highly successful at imputing and acquiring heterogeneous missing data and identifying outliers. However, within this specific application domain, existing VAE methods are restricted by using only one layer of latent variables and strictly Gaussian posterior approximations. To address these limitations, we present HH-VAEM, a Hierarchical VAE model for mixed-type incomplete data that uses Hamiltonian Monte Carlo with automatic hyper-parameter tuning for improved approximate inference. Our experiments show that HH-VAEM outperforms existing baselines in the tasks of missing data imputation, supervised learning and outlier identification with missing features. Finally, we also present a sampling-based approach for efficiently computing the information gain when missing features are to be acquired with HH-VAEM. Our experiments show that this sampling-based approach is superior to alternatives based on Gaussian approximations.
Addressing Bias in Active Learning with Depth Uncertainty Networks or Not
Dec 13, 2021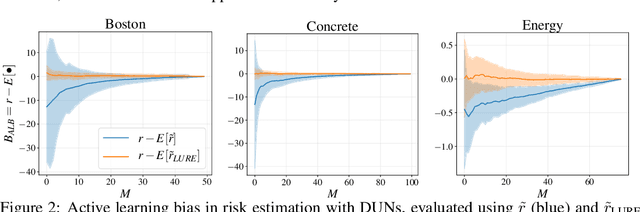
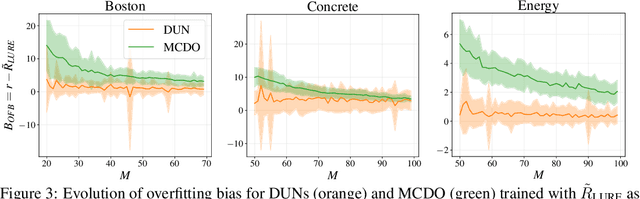
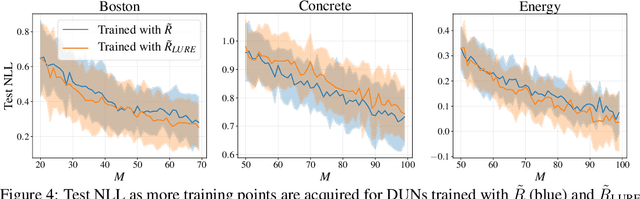
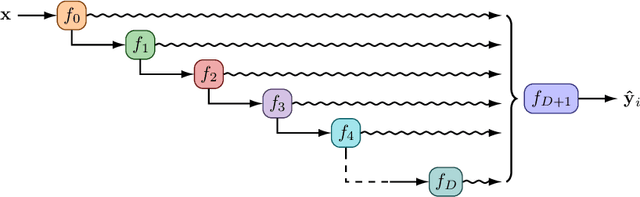
Abstract:Farquhar et al. [2021] show that correcting for active learning bias with underparameterised models leads to improved downstream performance. For overparameterised models such as NNs, however, correction leads either to decreased or unchanged performance. They suggest that this is due to an "overfitting bias" which offsets the active learning bias. We show that depth uncertainty networks operate in a low overfitting regime, much like underparameterised models. They should therefore see an increase in performance with bias correction. Surprisingly, they do not. We propose that this negative result, as well as the results Farquhar et al. [2021], can be explained via the lens of the bias-variance decomposition of generalisation error.
Depth Uncertainty Networks for Active Learning
Dec 13, 2021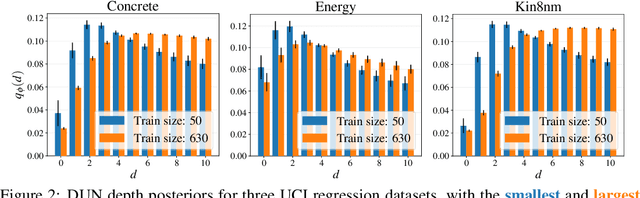
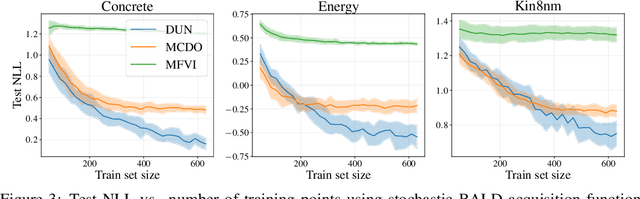
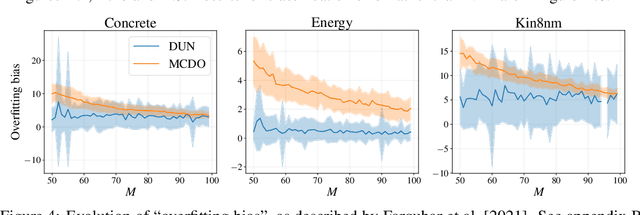
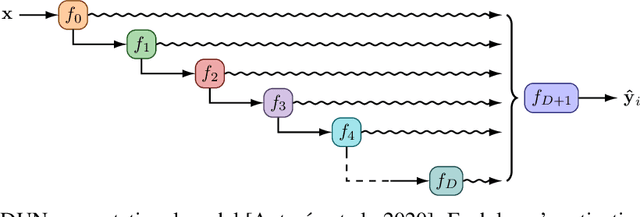
Abstract:In active learning, the size and complexity of the training dataset changes over time. Simple models that are well specified by the amount of data available at the start of active learning might suffer from bias as more points are actively sampled. Flexible models that might be well suited to the full dataset can suffer from overfitting towards the start of active learning. We tackle this problem using Depth Uncertainty Networks (DUNs), a BNN variant in which the depth of the network, and thus its complexity, is inferred. We find that DUNs outperform other BNN variants on several active learning tasks. Importantly, we show that on the tasks in which DUNs perform best they present notably less overfitting than baselines.
Bootstrap Your Flow
Dec 06, 2021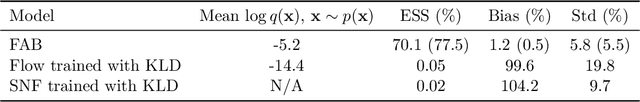
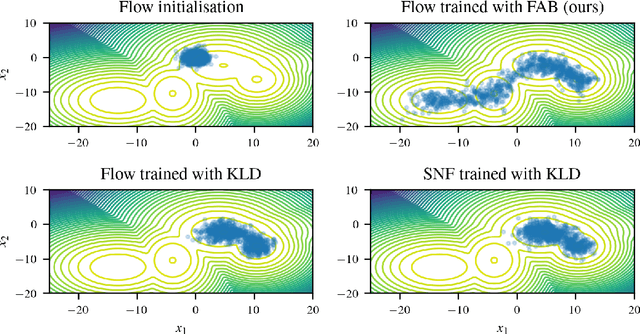
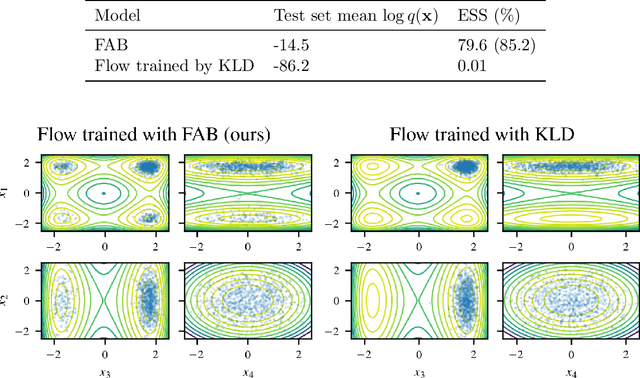
Abstract:Normalising flows are flexible, parameterized distributions that can be used to approximate expectations from intractable distributions via importance sampling. However, current flow-based approaches are limited on challenging targets where they either suffer from mode seeking behaviour or high variance in the training loss, or rely on samples from the target distribution, which may not be available. To address these challenges, we combine flows with annealed importance sampling (AIS), while using the $\alpha$-divergence as our objective, in a novel training procedure, FAB (Flow AIS Bootstrap). Thereby, the flow and AIS to improve each other in a bootstrapping manner. We demonstrate that FAB can be used to produce accurate approximations to complex target distributions, including Boltzmann distributions, in problems where previous flow-based methods fail.
Resampling Base Distributions of Normalizing Flows
Oct 29, 2021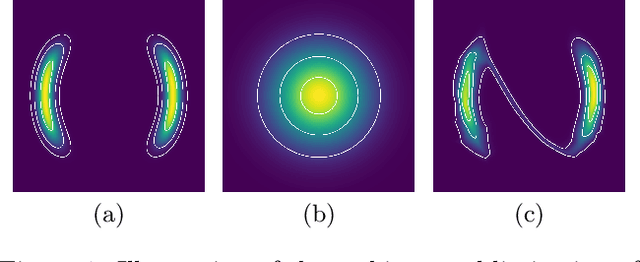
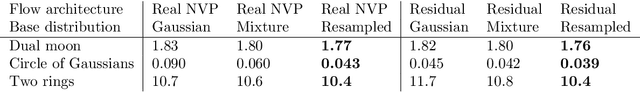
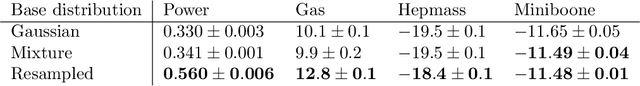
Abstract:Normalizing flows are a popular class of models for approximating probability distributions. However, their invertible nature limits their ability to model target distributions with a complex topological structure, such as Boltzmann distributions. Several procedures have been proposed to solve this problem but many of them sacrifice invertibility and, thereby, tractability of the log-likelihood as well as other desirable properties. To address these limitations, we introduce a base distribution for normalizing flows based on learned rejection sampling, allowing the resulting normalizing flow to model complex topologies without giving up bijectivity. Furthermore, we develop suitable learning algorithms using both maximizing the log-likelihood and the optimization of the reverse Kullback-Leibler divergence, and apply them to various sample problems, i.e.\ approximating 2D densities, density estimation of tabular data, image generation, and modeling Boltzmann distributions. In these experiments our method is competitive with or outperforms the baselines.
DOCKSTRING: easy molecular docking yields better benchmarks for ligand design
Oct 29, 2021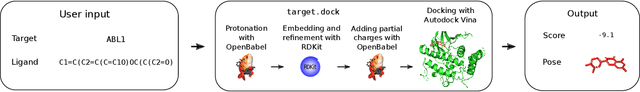
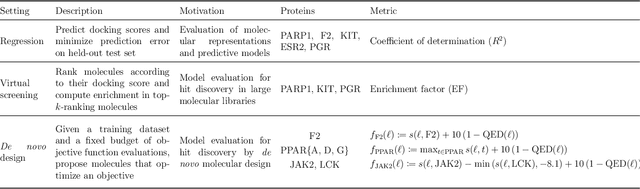
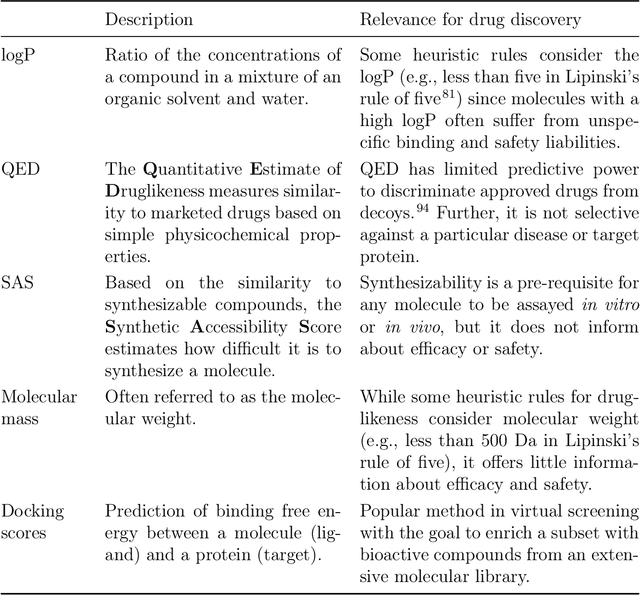
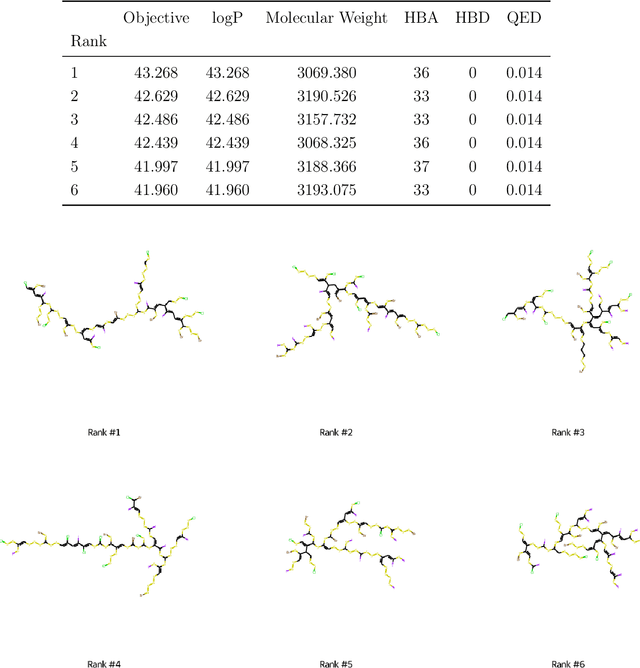
Abstract:The field of machine learning for drug discovery is witnessing an explosion of novel methods. These methods are often benchmarked on simple physicochemical properties such as solubility or general druglikeness, which can be readily computed. However, these properties are poor representatives of objective functions in drug design, mainly because they do not depend on the candidate's interaction with the target. By contrast, molecular docking is a widely successful method in drug discovery to estimate binding affinities. However, docking simulations require a significant amount of domain knowledge to set up correctly which hampers adoption. To this end, we present DOCKSTRING, a bundle for meaningful and robust comparison of ML models consisting of three components: (1) an open-source Python package for straightforward computation of docking scores; (2) an extensive dataset of docking scores and poses of more than 260K ligands for 58 medically-relevant targets; and (3) a set of pharmaceutically-relevant benchmark tasks including regression, virtual screening, and de novo design. The Python package implements a robust ligand and target preparation protocol that allows non-experts to obtain meaningful docking scores. Our dataset is the first to include docking poses, as well as the first of its size that is a full matrix, thus facilitating experiments in multiobjective optimization and transfer learning. Overall, our results indicate that docking scores are a more appropriate evaluation objective than simple physicochemical properties, yielding more realistic benchmark tasks and molecular candidates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge