Joakim Lindblad
From Cells to Survival: Hierarchical Analysis of Cell Inter-Relations in Multiplex Microscopy for Lung Cancer Prognosis
Dec 09, 2025Abstract:The tumor microenvironment (TME) has emerged as a promising source of prognostic biomarkers. To fully leverage its potential, analysis methods must capture complex interactions between different cell types. We propose HiGINE -- a hierarchical graph-based approach to predict patient survival (short vs. long) from TME characterization in multiplex immunofluorescence (mIF) images and enhance risk stratification in lung cancer. Our model encodes both local and global inter-relations in cell neighborhoods, incorporating information about cell types and morphology. Multimodal fusion, aggregating cancer stage with mIF-derived features, further boosts performance. We validate HiGINE on two public datasets, demonstrating improved risk stratification, robustness, and generalizability.
SLAM-AGS: Slide-Label Aware Multi-Task Pretraining Using Adaptive Gradient Surgery in Computational Cytology
Nov 18, 2025Abstract:Computational cytology faces two major challenges: i) instance-level labels are unreliable and prohibitively costly to obtain, ii) witness rates are extremely low. We propose SLAM-AGS, a Slide-Label-Aware Multitask pretraining framework that jointly optimizes (i) a weakly supervised similarity objective on slide-negative patches and (ii) a self-supervised contrastive objective on slide-positive patches, yielding stronger performance on downstream tasks. To stabilize learning, we apply Adaptive Gradient Surgery to tackle conflicting task gradients and prevent model collapse. We integrate the pretrained encoder into an attention-based Multiple Instance Learning aggregator for bag-level prediction and attention-guided retrieval of the most abnormal instances in a bag. On a publicly available bone-marrow cytology dataset, with simulated witness rates from 10% down to 0.5%, SLAM-AGS improves bag-level F1-Score and Top 400 positive cell retrieval over other pretraining methods, with the largest gains at low witness rates, showing that resolving gradient interference enables stable pretraining and better performance on downstream tasks. To facilitate reproducibility, we share our complete implementation and evaluation framework as open source: https://github.com/Ace95/SLAM-AGS.
Isolated Channel Vision Transformers: From Single-Channel Pretraining to Multi-Channel Finetuning
Mar 12, 2025Abstract:Vision Transformers (ViTs) have achieved remarkable success in standard RGB image processing tasks. However, applying ViTs to multi-channel imaging (MCI) data, e.g., for medical and remote sensing applications, remains a challenge. In particular, MCI data often consist of layers acquired from different modalities. Directly training ViTs on such data can obscure complementary information and impair the performance. In this paper, we introduce a simple yet effective pretraining framework for large-scale MCI datasets. Our method, named Isolated Channel ViT (IC-ViT), patchifies image channels individually and thereby enables pretraining for multimodal multi-channel tasks. We show that this channel-wise patchifying is a key technique for MCI processing. More importantly, one can pretrain the IC-ViT on single channels and finetune it on downstream multi-channel datasets. This pretraining framework captures dependencies between patches as well as channels and produces robust feature representation. Experiments on various tasks and benchmarks, including JUMP-CP and CHAMMI for cell microscopy imaging, and So2Sat-LCZ42 for satellite imaging, show that the proposed IC-ViT delivers 4-14 percentage points of performance improvement over existing channel-adaptive approaches. Further, its efficient training makes it a suitable candidate for large-scale pretraining of foundation models on heterogeneous data.
UpStory: the Uppsala Storytelling dataset
Jul 05, 2024Abstract:Friendship and rapport play an important role in the formation of constructive social interactions, and have been widely studied in educational settings due to their impact on student outcomes. Given the growing interest in automating the analysis of such phenomena through Machine Learning (ML), access to annotated interaction datasets is highly valuable. However, no dataset on dyadic child-child interactions explicitly capturing rapport currently exists. Moreover, despite advances in the automatic analysis of human behaviour, no previous work has addressed the prediction of rapport in child-child dyadic interactions in educational settings. We present UpStory -- the Uppsala Storytelling dataset: a novel dataset of naturalistic dyadic interactions between primary school aged children, with an experimental manipulation of rapport. Pairs of children aged 8-10 participate in a task-oriented activity: designing a story together, while being allowed free movement within the play area. We promote balanced collection of different levels of rapport by using a within-subjects design: self-reported friendships are used to pair each child twice, either minimizing or maximizing pair separation in the friendship network. The dataset contains data for 35 pairs, totalling 3h 40m of audio and video recordings. It includes two video sources covering the play area, as well as separate voice recordings for each child. An anonymized version of the dataset is made publicly available, containing per-frame head pose, body pose, and face features; as well as per-pair information, including the level of rapport. Finally, we provide ML baselines for the prediction of rapport.
Let it shine: Autofluorescence of Papanicolaou-stain improves AI-based cytological oral cancer detection
Jul 02, 2024



Abstract:Oral cancer is a global health challenge. It is treatable if detected early, but it is often fatal in late stages. There is a shift from the invasive and time-consuming tissue sampling and histological examination, toward non-invasive brush biopsies and cytological examination. Reliable computer-assisted methods are essential for cost-effective and accurate cytological analysis, but the lack of detailed cell-level annotations impairs model effectiveness. This study aims to improve AI-based oral cancer detection using multimodal imaging and deep fusion. We combine brightfield and fluorescence whole slide microscopy imaging to analyze Papanicolaou-stained liquid-based cytology slides of brush biopsies collected from both healthy and cancer patients. Due to limited cytological annotations, we utilize a weakly supervised deep learning approach using only patient-level labels. We evaluate various multimodal fusion strategies, including early, late, and three recent intermediate fusion methods. Our results show: (i) fluorescence imaging of Papanicolaou-stained samples provides substantial diagnostic information; (ii) multimodal fusion enhances classification and cancer detection accuracy over single-modality methods. Intermediate fusion is the leading method among the studied approaches. Specifically, the Co-Attention Fusion Network (CAFNet) model excels with an F1 score of 83.34% and accuracy of 91.79%, surpassing human performance on the task. Additional tests highlight the need for precise image registration to optimize multimodal analysis benefits. This study advances cytopathology by combining deep learning and multimodal imaging to enhance early, non-invasive detection of oral cancer, improving diagnostic accuracy and streamlining clinical workflows. The developed pipeline is also applicable in other cytological settings. Our codes and dataset are available online for further research.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Can representation learning for multimodal image registration be improved by supervision of intermediate layers?
Mar 01, 2023Abstract:Multimodal imaging and correlative analysis typically require image alignment. Contrastive learning can generate representations of multimodal images, reducing the challenging task of multimodal image registration to a monomodal one. Previously, additional supervision on intermediate layers in contrastive learning has improved biomedical image classification. We evaluate if a similar approach improves representations learned for registration to boost registration performance. We explore three approaches to add contrastive supervision to the latent features of the bottleneck layer in the U-Nets encoding the multimodal images and evaluate three different critic functions. Our results show that representations learned without additional supervision on latent features perform best in the downstream task of registration on two public biomedical datasets. We investigate the performance drop by exploiting recent insights in contrastive learning in classification and self-supervised learning. We visualize the spatial relations of the learned representations by means of multidimensional scaling, and show that additional supervision on the bottleneck layer can lead to partial dimensional collapse of the intermediate embedding space.
End-to-end Multiple Instance Learning with Gradient Accumulation
Mar 08, 2022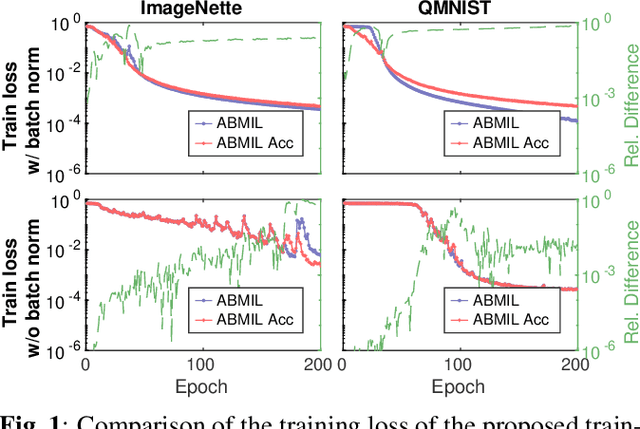
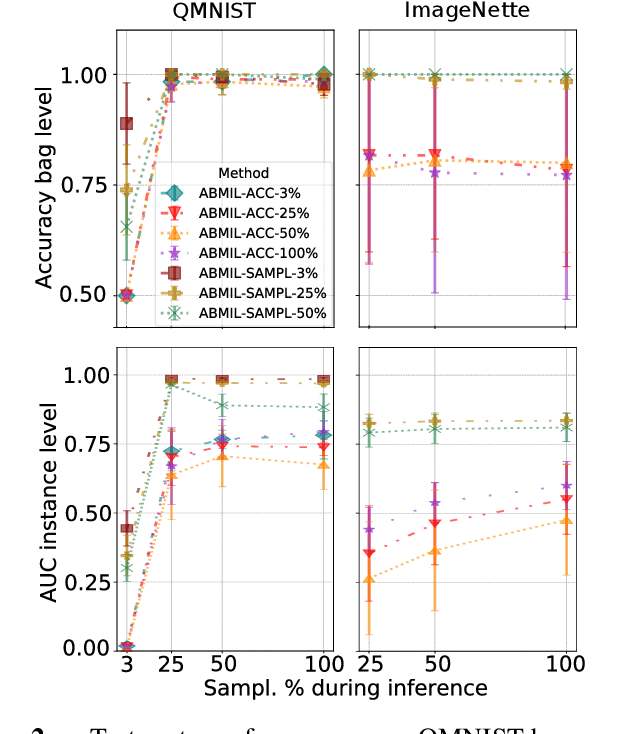
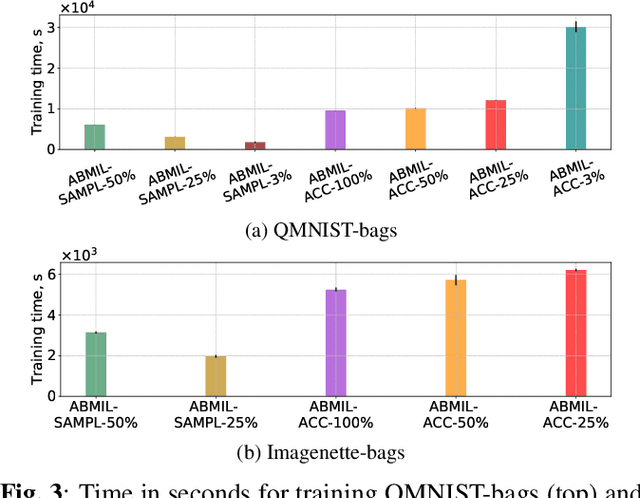
Abstract:Being able to learn on weakly labeled data, and provide interpretability, are two of the main reasons why attention-based deep multiple instance learning (ABMIL) methods have become particularly popular for classification of histopathological images. Such image data usually come in the form of gigapixel-sized whole-slide-images (WSI) that are cropped into smaller patches (instances). However, the sheer size of the data makes training of ABMIL models challenging. All the instances from one WSI cannot be processed at once by conventional GPUs. Existing solutions compromise training by relying on pre-trained models, strategic sampling or selection of instances, or self-supervised learning. We propose a training strategy based on gradient accumulation that enables direct end-to-end training of ABMIL models without being limited by GPU memory. We conduct experiments on both QMNIST and Imagenette to investigate the performance and training time, and compare with the conventional memory-expensive baseline and a recent sampled-based approach. This memory-efficient approach, although slower, reaches performance indistinguishable from the memory-expensive baseline.
Oral cancer detection and interpretation: Deep multiple instance learning versus conventional deep single instance learning
Feb 03, 2022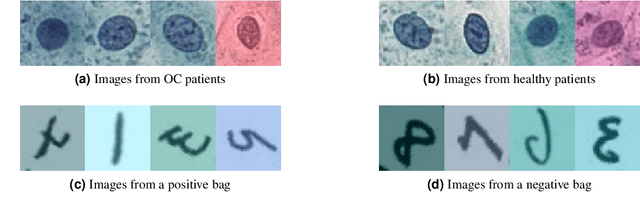
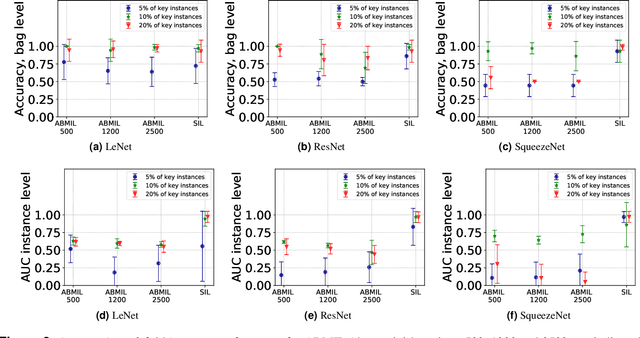
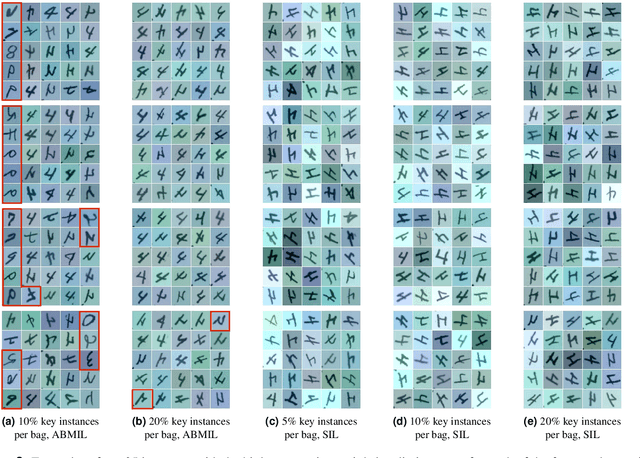
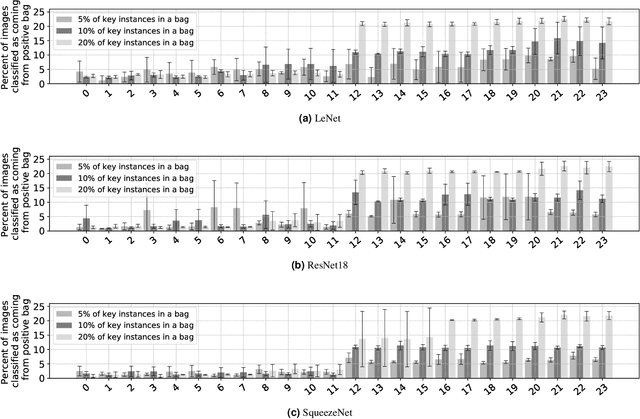
Abstract:The current medical standard for setting an oral cancer (OC) diagnosis is histological examination of a tissue sample from the oral cavity. This process is time consuming and more invasive than an alternative approach of acquiring a brush sample followed by cytological analysis. Skilled cytotechnologists are able to detect changes due to malignancy, however, to introduce this approach into clinical routine is associated with challenges such as a lack of experts and labour-intensive work. To design a trustworthy OC detection system that would assist cytotechnologists, we are interested in AI-based methods that reliably can detect cancer given only per-patient labels (minimizing annotation bias), and also provide information on which cells are most relevant for the diagnosis (enabling supervision and understanding). We, therefore, perform a comparison of a conventional single instance learning (SIL) approach and a modern multiple instance learning (MIL) method suitable for OC detection and interpretation, utilizing three different neural network architectures. To facilitate systematic evaluation of the considered approaches, we introduce a synthetic PAP-QMNIST dataset, that serves as a model of OC data, while offering access to per-instance ground truth. Our study indicates that on PAP-QMNIST, the SIL performs better, on average, than the MIL approach. Performance at the bag level on real-world cytological data is similar for both methods, yet the single instance approach performs better on average. Visual examination by cytotechnologist indicates that the methods manage to identify cells which deviate from normality, including malignant cells as well as those suspicious for dysplasia. We share the code as open source at https://github.com/MIDA-group/OralCancerMILvsSIL
Cross-Modality Sub-Image Retrieval using Contrastive Multimodal Image Representations
Jan 10, 2022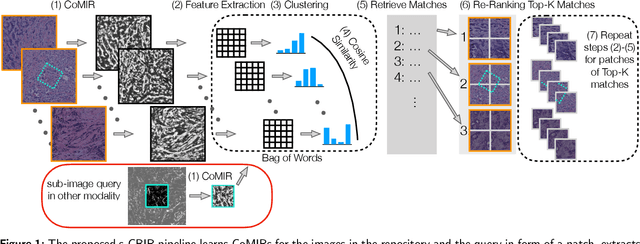
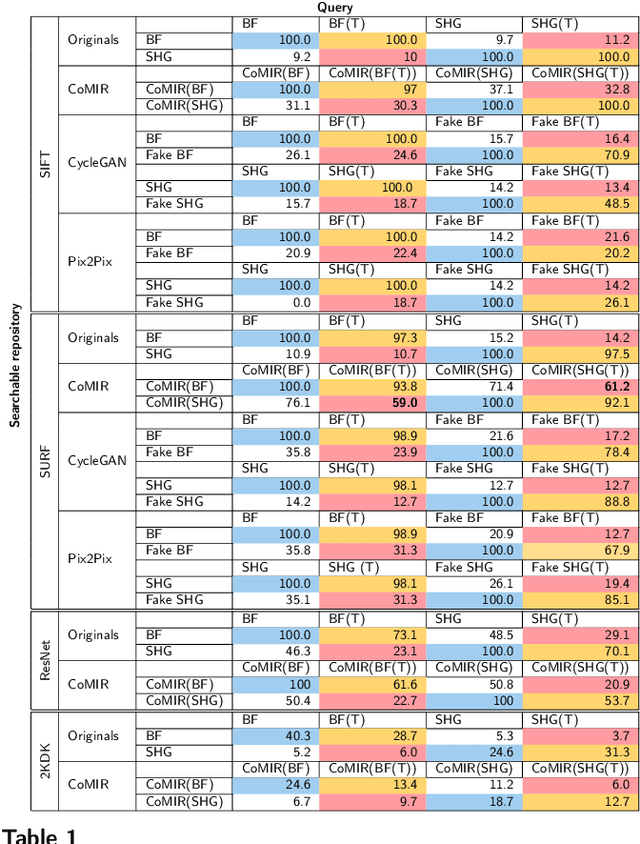
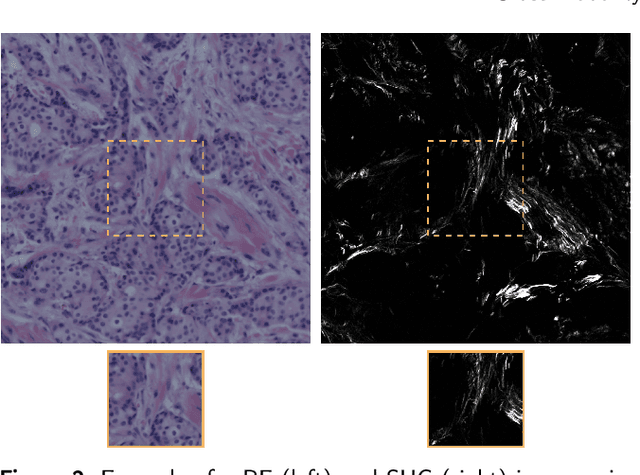
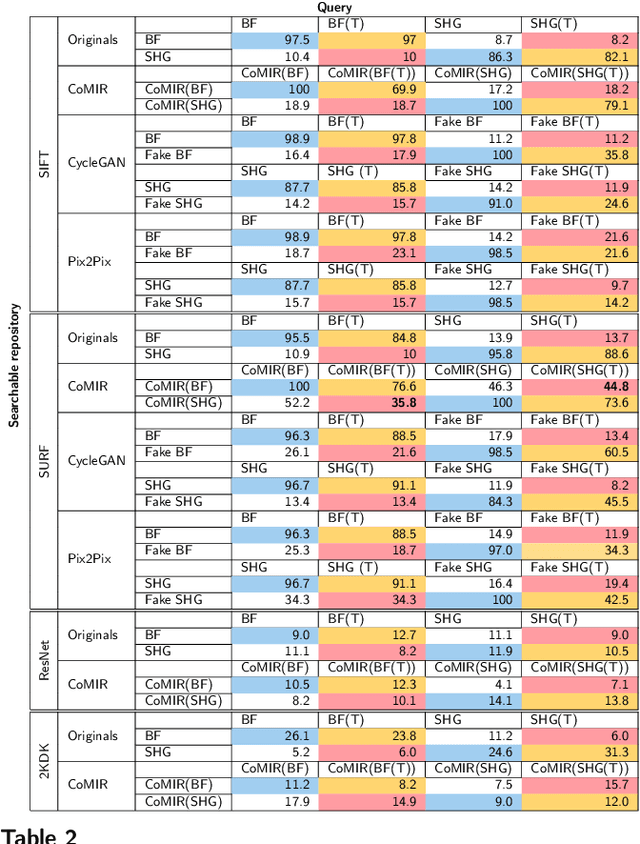
Abstract:In tissue characterization and cancer diagnostics, multimodal imaging has emerged as a powerful technique. Thanks to computational advances, large datasets can be exploited to improve diagnosis and discover patterns in pathologies. However, this requires efficient and scalable image retrieval methods. Cross-modality image retrieval is particularly demanding, as images of the same content captured in different modalities may display little common information. We propose a content-based image retrieval system (CBIR) for reverse (sub-)image search to retrieve microscopy images in one modality given a corresponding image captured by a different modality, where images are not aligned and share only few structures. We propose to combine deep learning to generate representations which embed both modalities in a common space, with classic, fast, and robust feature extractors (SIFT, SURF) to create a bag-of-words model for efficient and reliable retrieval. Our application-independent approach shows promising results on a publicly available dataset of brightfield and second harmonic generation microscopy images. We obtain 75.4% and 83.6% top-10 retrieval success for retrieval in one or the other direction. Our proposed method significantly outperforms both direct retrieval of the original multimodal (sub-)images, as well as their corresponding generative adversarial network (GAN)-based image-to-image translations. We establish that the proposed method performs better in comparison with a recent sub-image retrieval toolkit, GAN-based image-to-image translations, and learnt feature extractors for the downstream task of cross-modal image retrieval. We highlight the shortcomings of the latter methods and observe the importance of equivariance and invariance properties of the learnt representations and feature extractors in the CBIR pipeline. Code will be available at github.com/MIDA-group.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge