Jianxu Chen
Deep learning based Image Compression for Microscopy Images: An Empirical Study
Nov 02, 2023Abstract:With the fast development of modern microscopes and bioimaging techniques, an unprecedentedly large amount of imaging data are being generated, stored, analyzed, and even shared through networks. The size of the data poses great challenges for current data infrastructure. One common way to reduce the data size is by image compression. This present study analyzes classic and deep learning based image compression methods, and their impact on deep learning based image processing models. Deep learning based label-free prediction models (i.e., predicting fluorescent images from bright field images) are used as an example application for comparison and analysis. Effective image compression methods could help reduce the data size significantly without losing necessary information, and therefore reduce the burden on data management infrastructure and permit fast transmission through the network for data sharing or cloud computing. To compress images in such a wanted way, multiple classical lossy image compression techniques are compared to several AI-based compression models provided by and trained with the CompressAI toolbox using python. These different compression techniques are compared in compression ratio, multiple image similarity measures and, most importantly, the prediction accuracy from label-free models on compressed images. We found that AI-based compression techniques largely outperform the classic ones and will minimally affect the downstream label-free task in 2D cases. In the end, we hope the present study could shed light on the potential of deep learning based image compression and the impact of image compression on downstream deep learning based image analysis models.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Masked Diffusion as Self-supervised Representation Learner
Aug 27, 2023Abstract:Denoising diffusion probabilistic models have recently demonstrated state-of-the-art generative performance and been used as strong pixel-level representation learners. This paper decomposes the interrelation between the generative capability and representation learning ability inherent in diffusion models. We present masked diffusion model (MDM), a scalable self-supervised representation learner that substitutes the conventional additive Gaussian noise of traditional diffusion with a masking mechanism. Our proposed approach convincingly surpasses prior benchmarks, demonstrating remarkable advancements in both medical and natural image semantic segmentation tasks, particularly within the context of few-shot scenario.
EfficientBioAI: Making Bioimaging AI Models Efficient in Energy, Latency and Representation
Jun 09, 2023Abstract:Artificial intelligence (AI) has been widely used in bioimage image analysis nowadays, but the efficiency of AI models, like the energy consumption and latency is not ignorable due to the growing model size and complexity, as well as the fast-growing analysis needs in modern biomedical studies. Like we can compress large images for efficient storage and sharing, we can also compress the AI models for efficient applications and deployment. In this work, we present EfficientBioAI, a plug-and-play toolbox that can compress given bioimaging AI models for them to run with significantly reduced energy cost and inference time on both CPU and GPU, without compromise on accuracy. In some cases, the prediction accuracy could even increase after compression, since the compression procedure could remove redundant information in the model representation and therefore reduce over-fitting. From four different bioimage analysis applications, we observed around 2-5 times speed-up during inference and 30-80$\%$ saving in energy. Cutting the runtime of large scale bioimage analysis from days to hours or getting a two-minutes bioimaging AI model inference done in near real-time will open new doors for method development and biomedical discoveries. We hope our toolbox will facilitate resource-constrained bioimaging AI and accelerate large-scale AI-based quantitative biological studies in an eco-friendly way, as well as stimulate further research on the efficiency of bioimaging AI.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
MMV_Im2Im: An Open Source Microscopy Machine Vision Toolbox for Image-to-Image Transformation
Sep 06, 2022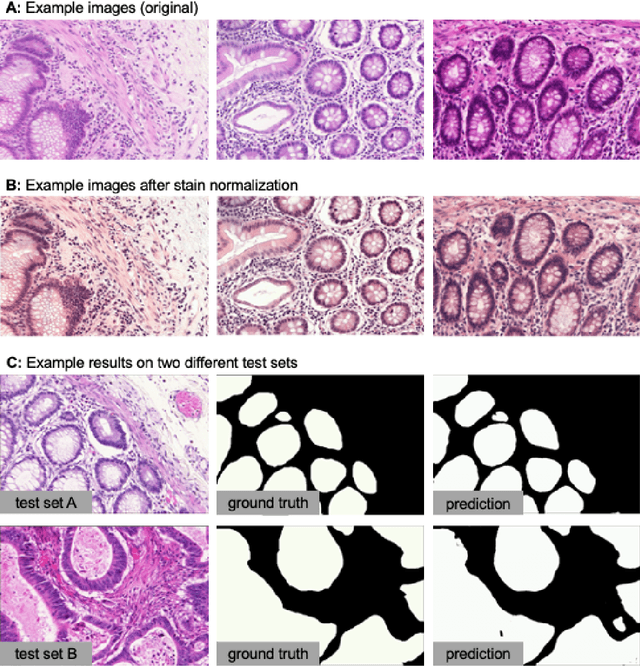
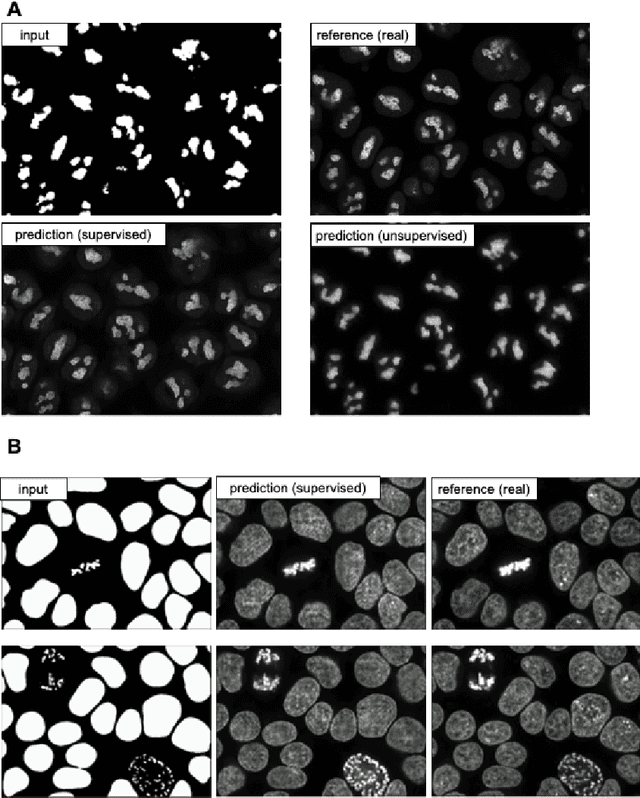
Abstract:The deep learning research in computer vision has been growing extremely fast in the past decade, many of which have been translated into novel image analysis methods for biomedical problems. Broadly speaking, many deep learning based biomedical image analysis methods can be considered as a general image-to-image transformation framework. In this work, we introduce a new open source python package MMV_Im2Im for image-to-image transformation in bioimaging applications. The overall package is designed with a generic image-to-image transformation framework, which could be directly used for semantic segmentation, instance segmentation, image restoration, image generation, etc.. The implementation takes advantage of the state-of-the-art machine learning engineering techniques for users to focus on the research without worrying about the engineering details. We demonstrate the effectiveness of MMV_Im2Im in more than ten different biomedical problems. For biomedical machine learning researchers, we hope this new package could serve as the starting point for their specific problems to stimulate new biomedical image analysis or machine learning methods. For experimental biomedical researchers, we hope this work can provide a holistic view of the image-to-image transformation concept with diverse examples, so that deep learning based image-to-image transformation could be further integrated into the assay development process and permit new biomedical studies that can hardly be done only with traditional experimental methods. Source code can be found at https://github.com/MMV-Lab/mmv_im2im.
H-EMD: A Hierarchical Earth Mover's Distance Method for Instance Segmentation
Jun 02, 2022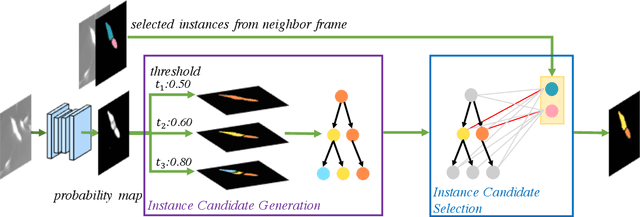
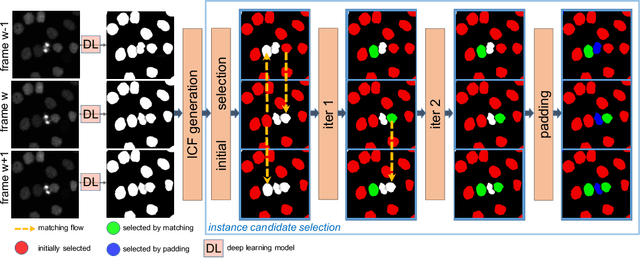
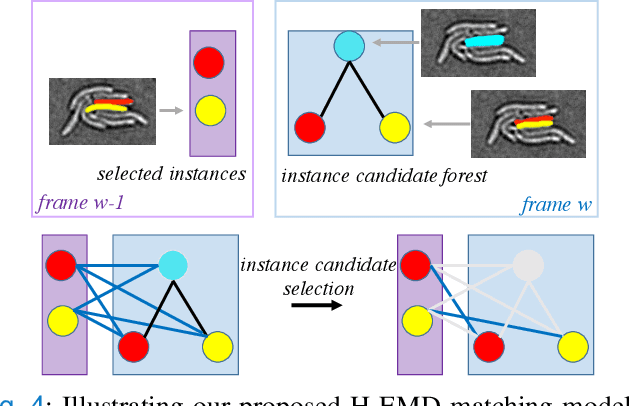

Abstract:Deep learning (DL) based semantic segmentation methods have achieved excellent performance in biomedical image segmentation, producing high quality probability maps to allow extraction of rich instance information to facilitate good instance segmentation. While numerous efforts were put into developing new DL semantic segmentation models, less attention was paid to a key issue of how to effectively explore their probability maps to attain the best possible instance segmentation. We observe that probability maps by DL semantic segmentation models can be used to generate many possible instance candidates, and accurate instance segmentation can be achieved by selecting from them a set of "optimized" candidates as output instances. Further, the generated instance candidates form a well-behaved hierarchical structure (a forest), which allows selecting instances in an optimized manner. Hence, we propose a novel framework, called hierarchical earth mover's distance (H-EMD), for instance segmentation in biomedical 2D+time videos and 3D images, which judiciously incorporates consistent instance selection with semantic-segmentation-generated probability maps. H-EMD contains two main stages. (1) Instance candidate generation: capturing instance-structured information in probability maps by generating many instance candidates in a forest structure. (2) Instance candidate selection: selecting instances from the candidate set for final instance segmentation. We formulate a key instance selection problem on the instance candidate forest as an optimization problem based on the earth mover's distance (EMD), and solve it by integer linear programming. Extensive experiments on eight biomedical video or 3D datasets demonstrate that H-EMD consistently boosts DL semantic segmentation models and is highly competitive with state-of-the-art methods.
Unlabeled Data Guided Semi-supervised Histopathology Image Segmentation
Dec 17, 2020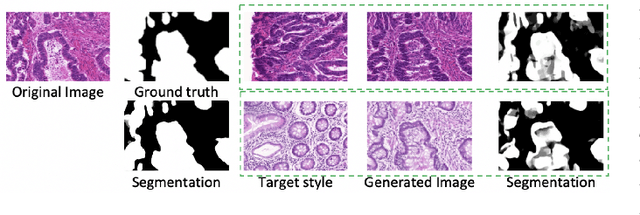
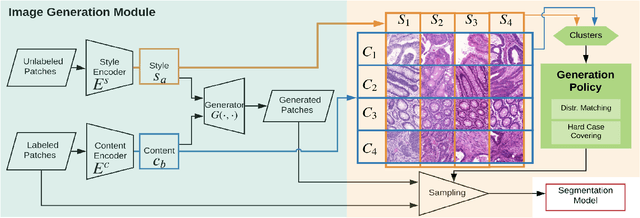
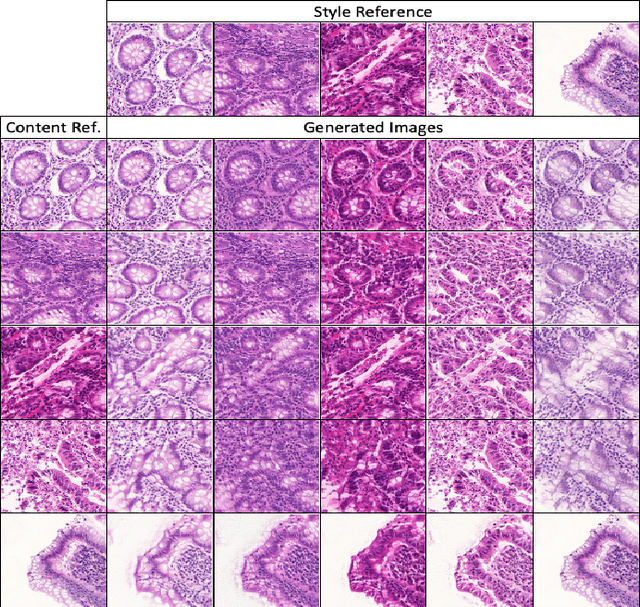
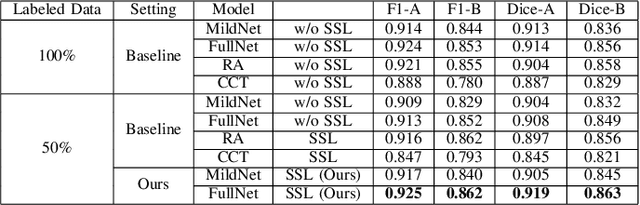
Abstract:Automatic histopathology image segmentation is crucial to disease analysis. Limited available labeled data hinders the generalizability of trained models under the fully supervised setting. Semi-supervised learning (SSL) based on generative methods has been proven to be effective in utilizing diverse image characteristics. However, it has not been well explored what kinds of generated images would be more useful for model training and how to use such images. In this paper, we propose a new data guided generative method for histopathology image segmentation by leveraging the unlabeled data distributions. First, we design an image generation module. Image content and style are disentangled and embedded in a clustering-friendly space to utilize their distributions. New images are synthesized by sampling and cross-combining contents and styles. Second, we devise an effective data selection policy for judiciously sampling the generated images: (1) to make the generated training set better cover the dataset, the clusters that are underrepresented in the original training set are covered more; (2) to make the training process more effective, we identify and oversample the images of "hard cases" in the data for which annotated training data may be scarce. Our method is evaluated on glands and nuclei datasets. We show that under both the inductive and transductive settings, our SSL method consistently boosts the performance of common segmentation models and attains state-of-the-art results.
Cascade Decoder: A Universal Decoding Method for Biomedical Image Segmentation
Jan 15, 2019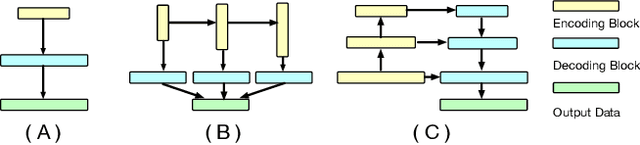
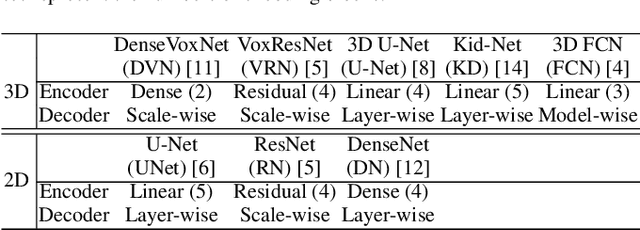
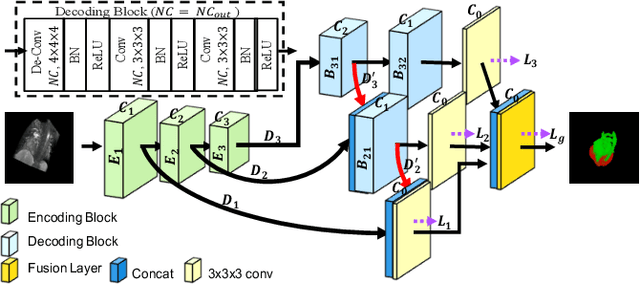
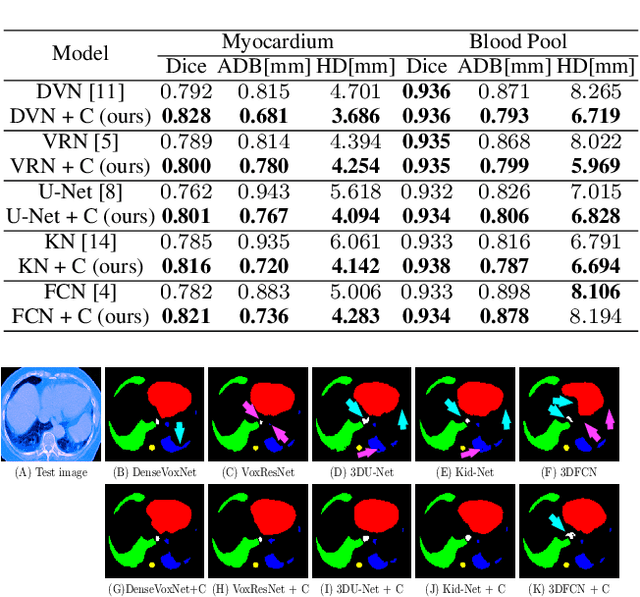
Abstract:The Encoder-Decoder architecture is a main stream deep learning model for biomedical image segmentation. The encoder fully compresses the input and generates encoded features, and the decoder then produces dense predictions using encoded features. However, decoders are still under-explored in such architectures. In this paper, we comprehensively study the state-of-the-art Encoder-Decoder architectures, and propose a new universal decoder, called cascade decoder, to improve semantic segmentation accuracy. Our cascade decoder can be embedded into existing networks and trained altogether in an end-to-end fashion. The cascade decoder structure aims to conduct more effective decoding of hierarchically encoded features and is more compatible with common encoders than the known decoders. We replace the decoders of state-of-the-art models with our cascade decoder for several challenging biomedical image segmentation tasks, and the considerable improvements achieved demonstrate the efficacy of our new decoding method.
A New Registration Approach for Dynamic Analysis of Calcium Signals in Organs
Feb 01, 2018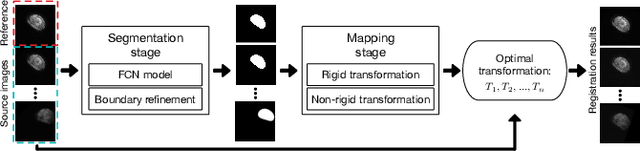
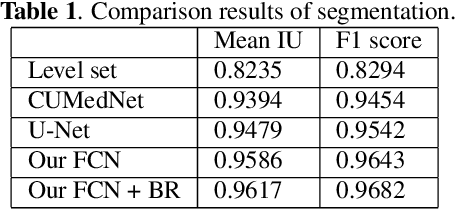
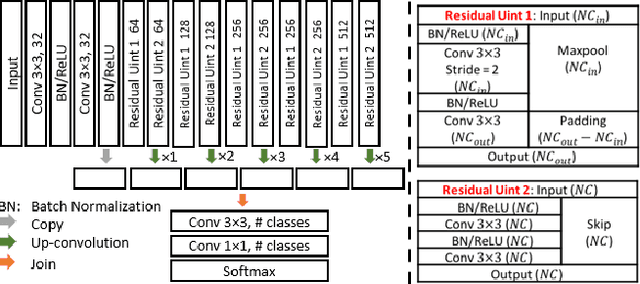
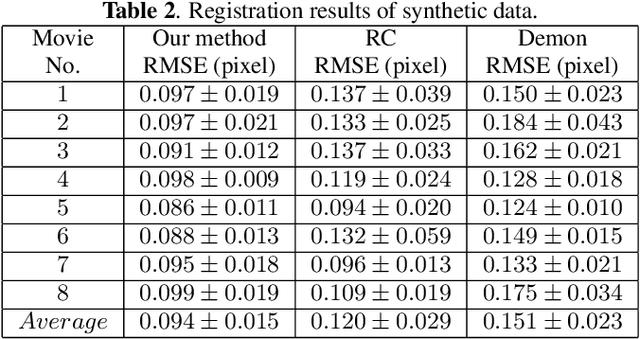
Abstract:Wing disc pouches of fruit flies are a powerful genetic model for studying physiological intercellular calcium ($Ca^{2+}$) signals for dynamic analysis of cell signaling in organ development and disease studies. A key to analyzing spatial-temporal patterns of $Ca^{2+}$ signal waves is to accurately align the pouches across image sequences. However, pouches in different image frames may exhibit extensive intensity oscillations due to $Ca^{2+}$ signaling dynamics, and commonly used multimodal non-rigid registration methods may fail to achieve satisfactory results. In this paper, we develop a new two-phase non-rigid registration approach to register pouches in image sequences. First, we conduct segmentation of the region of interest. (i.e., pouches) using a deep neural network model. Second, we obtain an optimal transformation and align pouches across the image sequences. Evaluated using both synthetic data and real pouch data, our method considerably outperforms the state-of-the-art non-rigid registration methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge