Jesus Pineda
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Corneal endothelium assessment in specular microscopy images with Fuchs' dystrophy via deep regression of signed distance maps
Oct 13, 2022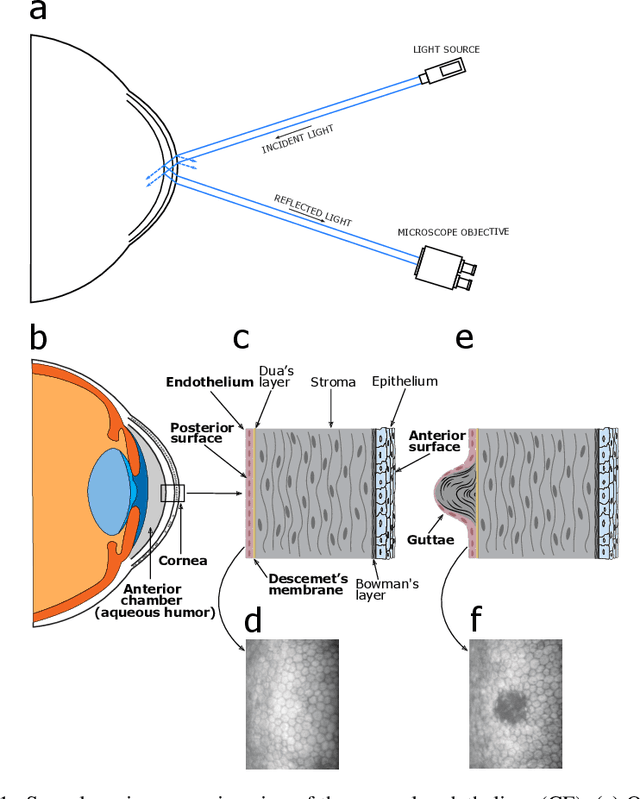
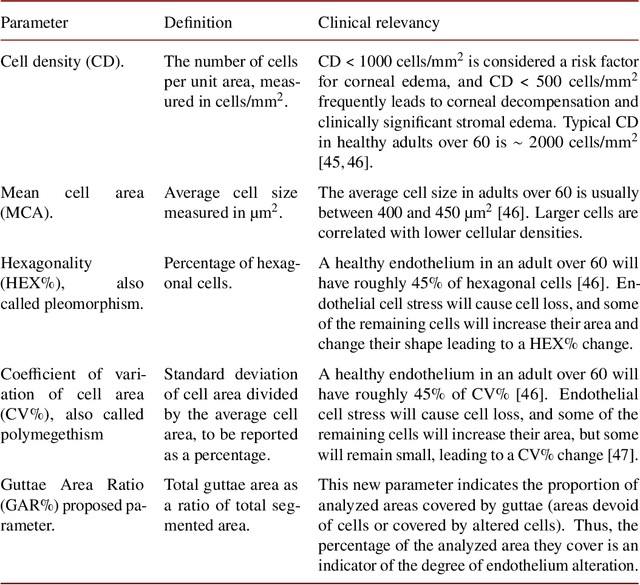
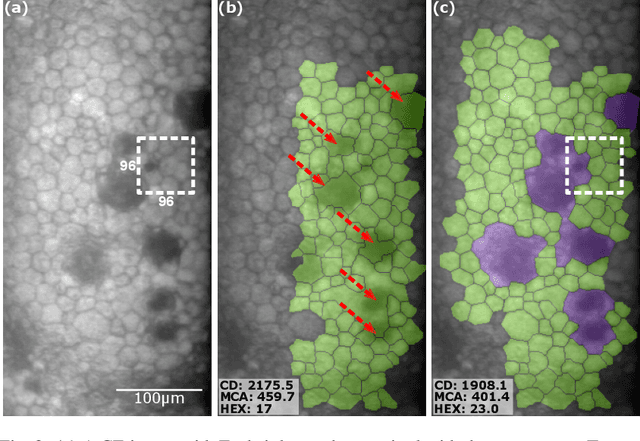
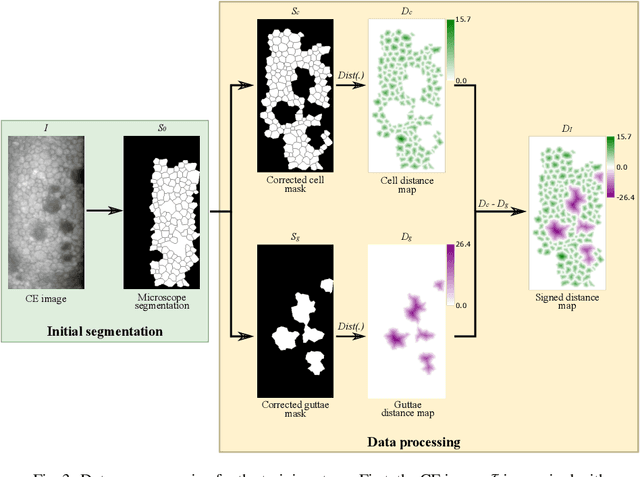
Abstract:Specular microscopy assessment of the human corneal endothelium (CE) in Fuchs' dystrophy is challenging due to the presence of dark image regions called guttae. This paper proposes a UNet-based segmentation approach that requires minimal post-processing and achieves reliable CE morphometric assessment and guttae identification across all degrees of Fuchs' dystrophy. We cast the segmentation problem as a regression task of the cell and gutta signed distance maps instead of a pixel-level classification task as typically done with UNets. Compared to the conventional UNet classification approach, the distance-map regression approach converges faster in clinically relevant parameters. It also produces morphometric parameters that agree with the manually-segmented ground-truth data, namely the average cell density difference of -41.9 cells/mm2 (95% confidence interval (CI) [-306.2, 222.5]) and the average difference of mean cell area of 14.8 um2 (95% CI [-41.9, 71.5]). These results suggest a promising alternative for CE assessment.
Anomaly Detection in Unstructured Environments using Bayesian Nonparametric Scene Modeling
Feb 16, 2016
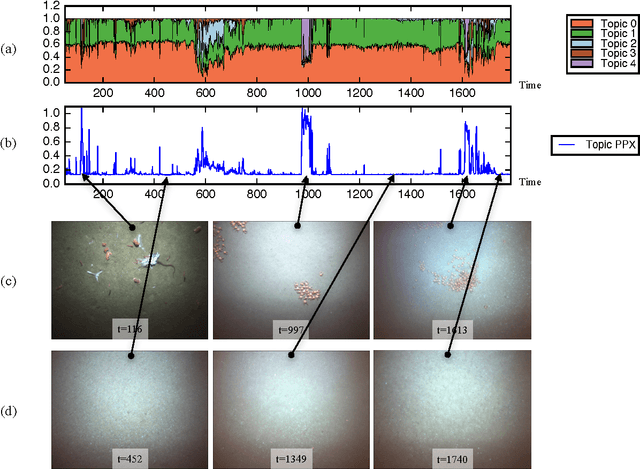
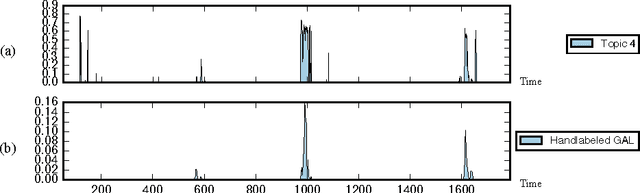
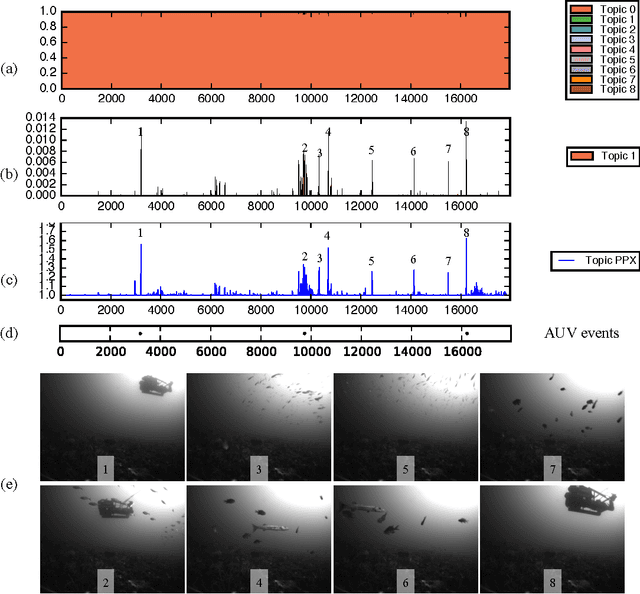
Abstract:This paper explores the use of a Bayesian non-parametric topic modeling technique for the purpose of anomaly detection in video data. We present results from two experiments. The first experiment shows that the proposed technique is automatically able characterize the underlying terrain, and detect anomalous flora in image data collected by an underwater robot. The second experiment shows that the same technique can be used on images from a static camera in a dynamic unstructured environment. In the second dataset, consisting of video data from a static seafloor camera capturing images of a busy coral reef, the proposed technique was able to detect all three instances of an underwater vehicle passing in front of the camera, amongst many other observations of fishes, debris, lighting changes due to surface waves, and benthic flora.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge