Jan Egger
Classification of lung cancer subtypes on CT images with synthetic pathological priors
Aug 09, 2023



Abstract:The accurate diagnosis on pathological subtypes for lung cancer is of significant importance for the follow-up treatments and prognosis managements. In this paper, we propose self-generating hybrid feature network (SGHF-Net) for accurately classifying lung cancer subtypes on computed tomography (CT) images. Inspired by studies stating that cross-scale associations exist in the image patterns between the same case's CT images and its pathological images, we innovatively developed a pathological feature synthetic module (PFSM), which quantitatively maps cross-modality associations through deep neural networks, to derive the "gold standard" information contained in the corresponding pathological images from CT images. Additionally, we designed a radiological feature extraction module (RFEM) to directly acquire CT image information and integrated it with the pathological priors under an effective feature fusion framework, enabling the entire classification model to generate more indicative and specific pathologically related features and eventually output more accurate predictions. The superiority of the proposed model lies in its ability to self-generate hybrid features that contain multi-modality image information based on a single-modality input. To evaluate the effectiveness, adaptability, and generalization ability of our model, we performed extensive experiments on a large-scale multi-center dataset (i.e., 829 cases from three hospitals) to compare our model and a series of state-of-the-art (SOTA) classification models. The experimental results demonstrated the superiority of our model for lung cancer subtypes classification with significant accuracy improvements in terms of accuracy (ACC), area under the curve (AUC), and F1 score.
FAM: Relative Flatness Aware Minimization
Jul 05, 2023



Abstract:Flatness of the loss curve around a model at hand has been shown to empirically correlate with its generalization ability. Optimizing for flatness has been proposed as early as 1994 by Hochreiter and Schmidthuber, and was followed by more recent successful sharpness-aware optimization techniques. Their widespread adoption in practice, though, is dubious because of the lack of theoretically grounded connection between flatness and generalization, in particular in light of the reparameterization curse - certain reparameterizations of a neural network change most flatness measures but do not change generalization. Recent theoretical work suggests that a particular relative flatness measure can be connected to generalization and solves the reparameterization curse. In this paper, we derive a regularizer based on this relative flatness that is easy to compute, fast, efficient, and works with arbitrary loss functions. It requires computing the Hessian only of a single layer of the network, which makes it applicable to large neural networks, and with it avoids an expensive mapping of the loss surface in the vicinity of the model. In an extensive empirical evaluation we show that this relative flatness aware minimization (FAM) improves generalization in a multitude of applications and models, both in finetuning and standard training. We make the code available at github.
Why does my medical AI look at pictures of birds? Exploring the efficacy of transfer learning across domain boundaries
Jun 30, 2023Abstract:It is an open secret that ImageNet is treated as the panacea of pretraining. Particularly in medical machine learning, models not trained from scratch are often finetuned based on ImageNet-pretrained models. We posit that pretraining on data from the domain of the downstream task should almost always be preferred instead. We leverage RadNet-12M, a dataset containing more than 12 million computed tomography (CT) image slices, to explore the efficacy of self-supervised pretraining on medical and natural images. Our experiments cover intra- and cross-domain transfer scenarios, varying data scales, finetuning vs. linear evaluation, and feature space analysis. We observe that intra-domain transfer compares favorably to cross-domain transfer, achieving comparable or improved performance (0.44% - 2.07% performance increase using RadNet pretraining, depending on the experiment) and demonstrate the existence of a domain boundary-related generalization gap and domain-specific learned features.
CellViT: Vision Transformers for Precise Cell Segmentation and Classification
Jun 27, 2023



Abstract:Nuclei detection and segmentation in hematoxylin and eosin-stained (H&E) tissue images are important clinical tasks and crucial for a wide range of applications. However, it is a challenging task due to nuclei variances in staining and size, overlapping boundaries, and nuclei clustering. While convolutional neural networks have been extensively used for this task, we explore the potential of Transformer-based networks in this domain. Therefore, we introduce a new method for automated instance segmentation of cell nuclei in digitized tissue samples using a deep learning architecture based on Vision Transformer called CellViT. CellViT is trained and evaluated on the PanNuke dataset, which is one of the most challenging nuclei instance segmentation datasets, consisting of nearly 200,000 annotated Nuclei into 5 clinically important classes in 19 tissue types. We demonstrate the superiority of large-scale in-domain and out-of-domain pre-trained Vision Transformers by leveraging the recently published Segment Anything Model and a ViT-encoder pre-trained on 104 million histological image patches - achieving state-of-the-art nuclei detection and instance segmentation performance on the PanNuke dataset with a mean panoptic quality of 0.51 and an F1-detection score of 0.83. The code is publicly available at https://github.com/TIO-IKIM/CellViT
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Open-Source Skull Reconstruction with MONAI
Nov 25, 2022



Abstract:We present a deep learning-based approach for skull reconstruction for MONAI, which has been pre-trained on the MUG500+ skull dataset. The implementation follows the MONAI contribution guidelines, hence, it can be easily tried out and used, and extended by MONAI users. The primary goal of this paper lies in the investigation of open-sourcing codes and pre-trained deep learning models under the MONAI framework. Nowadays, open-sourcing software, especially (pre-trained) deep learning models, has become increasingly important. Over the years, medical image analysis experienced a tremendous transformation. Over a decade ago, algorithms had to be implemented and optimized with low-level programming languages, like C or C++, to run in a reasonable time on a desktop PC, which was not as powerful as today's computers. Nowadays, users have high-level scripting languages like Python, and frameworks like PyTorch and TensorFlow, along with a sea of public code repositories at hand. As a result, implementations that had thousands of lines of C or C++ code in the past, can now be scripted with a few lines and in addition executed in a fraction of the time. To put this even on a higher level, the Medical Open Network for Artificial Intelligence (MONAI) framework tailors medical imaging research to an even more convenient process, which can boost and push the whole field. The MONAI framework is a freely available, community-supported, open-source and PyTorch-based framework, that also enables to provide research contributions with pre-trained models to others. Codes and pre-trained weights for skull reconstruction are publicly available at: https://github.com/Project-MONAI/research-contributions/tree/master/SkullRec
'A net for everyone': fully personalized and unsupervised neural networks trained with longitudinal data from a single patient
Oct 25, 2022Abstract:With the rise in importance of personalized medicine, we trained personalized neural networks to detect tumor progression in longitudinal datasets. The model was evaluated on two datasets with a total of 64 scans from 32 patients diagnosed with glioblastoma multiforme (GBM). Contrast-enhanced T1w sequences of brain magnetic resonance imaging (MRI) images were used in this study. For each patient, we trained their own neural network using just two images from different timepoints. Our approach uses a Wasserstein-GAN (generative adversarial network), an unsupervised network architecture, to map the differences between the two images. Using this map, the change in tumor volume can be evaluated. Due to the combination of data augmentation and the network architecture, co-registration of the two images is not needed. Furthermore, we do not rely on any additional training data, (manual) annotations or pre-training neural networks. The model received an AUC-score of 0.87 for tumor change. We also introduced a modified RANO criteria, for which an accuracy of 66% can be achieved. We show that using data from just one patient can be used to train deep neural networks to monitor tumor change.
Training β-VAE by Aggregating a Learned Gaussian Posterior with a Decoupled Decoder
Sep 29, 2022
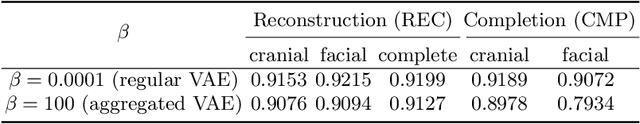
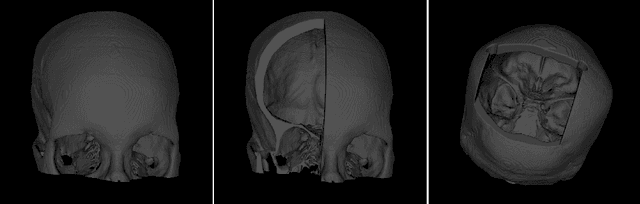
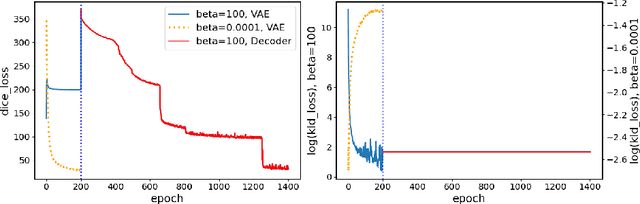
Abstract:The reconstruction loss and the Kullback-Leibler divergence (KLD) loss in a variational autoencoder (VAE) often play antagonistic roles, and tuning the weight of the KLD loss in $\beta$-VAE to achieve a balance between the two losses is a tricky and dataset-specific task. As a result, current practices in VAE training often result in a trade-off between the reconstruction fidelity and the continuity$/$disentanglement of the latent space, if the weight $\beta$ is not carefully tuned. In this paper, we present intuitions and a careful analysis of the antagonistic mechanism of the two losses, and propose, based on the insights, a simple yet effective two-stage method for training a VAE. Specifically, the method aggregates a learned Gaussian posterior $z \sim q_{\theta} (z|x)$ with a decoder decoupled from the KLD loss, which is trained to learn a new conditional distribution $p_{\phi} (x|z)$ of the input data $x$. Experimentally, we show that the aggregated VAE maximally satisfies the Gaussian assumption about the latent space, while still achieves a reconstruction error comparable to when the latent space is only loosely regularized by $\mathcal{N}(\mathbf{0},I)$. The proposed approach does not require hyperparameter (i.e., the KLD weight $\beta$) tuning given a specific dataset as required in common VAE training practices. We evaluate the method using a medical dataset intended for 3D skull reconstruction and shape completion, and the results indicate promising generative capabilities of the VAE trained using the proposed method. Besides, through guided manipulation of the latent variables, we establish a connection between existing autoencoder (AE)-based approaches and generative approaches, such as VAE, for the shape completion problem. Codes and pre-trained weights are available at https://github.com/Jianningli/skullVAE
The HoloLens in Medicine: A systematic Review and Taxonomy
Sep 06, 2022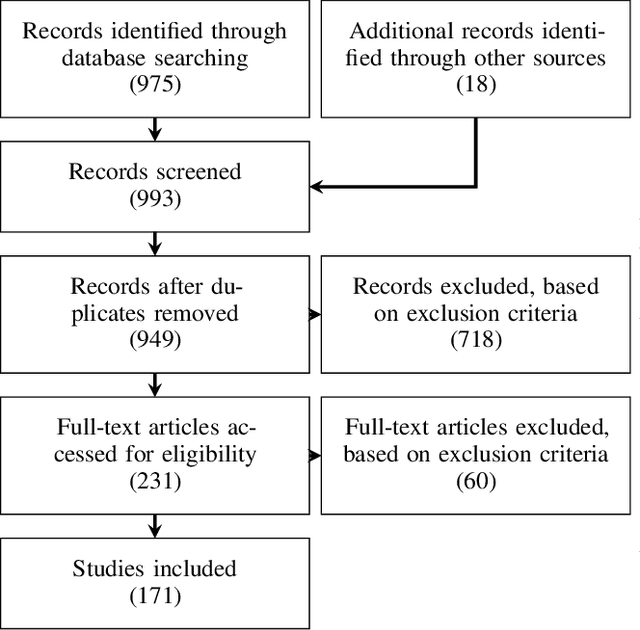
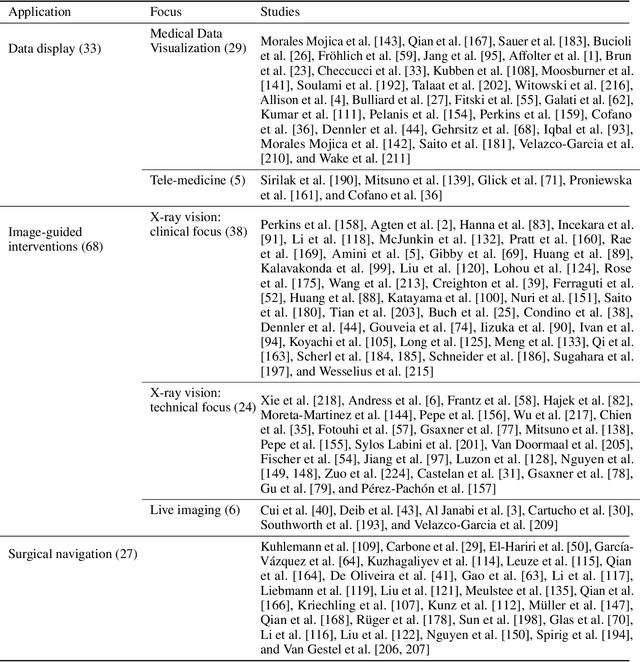
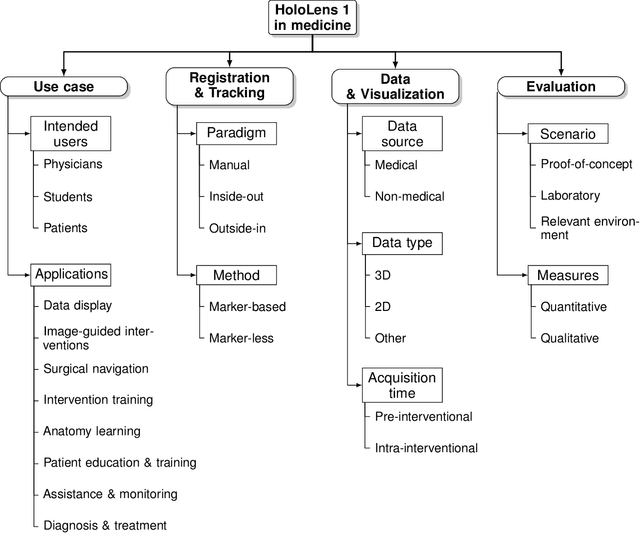

Abstract:The HoloLens (Microsoft Corp., Redmond, WA), a head-worn, optically see-through augmented reality display, is the main player in the recent boost in medical augmented reality research. In medical settings, the HoloLens enables the physician to obtain immediate insight into patient information, directly overlaid with their view of the clinical scenario, the medical student to gain a better understanding of complex anatomies or procedures, and even the patient to execute therapeutic tasks with improved, immersive guidance. In this systematic review, we provide a comprehensive overview of the usage of the first-generation HoloLens within the medical domain, from its release in March 2016, until the year of 2021, were attention is shifting towards it's successor, the HoloLens 2. We identified 171 relevant publications through a systematic search of the PubMed and Scopus databases. We analyze these publications in regard to their intended use case, technical methodology for registration and tracking, data sources, visualization as well as validation and evaluation. We find that, although the feasibility of using the HoloLens in various medical scenarios has been shown, increased efforts in the areas of precision, reliability, usability, workflow and perception are necessary to establish AR in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge