Ida-Maria Sintorn
PARIC: Probabilistic Attention Regularization for Language Guided Image Classification from Pre-trained Vison Language Models
Mar 14, 2025



Abstract:Language-guided attention frameworks have significantly enhanced both interpretability and performance in image classification; however, the reliance on deterministic embeddings from pre-trained vision-language foundation models to generate reference attention maps frequently overlooks the intrinsic multivaluedness and ill-posed characteristics of cross-modal mappings. To address these limitations, we introduce PARIC, a probabilistic framework for guiding visual attention via language specifications. Our approach enables pre-trained vision-language models to generate probabilistic reference attention maps, which align textual and visual modalities more effectively while incorporating uncertainty estimates, as compared to their deterministic counterparts. Experiments on benchmark test problems demonstrate that PARIC enhances prediction accuracy, mitigates bias, ensures consistent predictions, and improves robustness across various datasets.
Efficient High-Resolution Template Matching with Vector Quantized Nearest Neighbour Fields
Jun 26, 2023Abstract:Template matching is a fundamental problem in computer vision and has applications in various fields, such as object detection, image registration, and object tracking. The current state-of-the-art methods rely on nearest-neighbour (NN) matching in which the query feature space is converted to NN space by representing each query pixel with its NN in the template pixels. The NN-based methods have been shown to perform better in occlusions, changes in appearance, illumination variations, and non-rigid transformations. However, NN matching scales poorly with high-resolution data and high feature dimensions. In this work, we present an NN-based template-matching method which efficiently reduces the NN computations and introduces filtering in the NN fields to consider deformations. A vector quantization step first represents the template with $k$ features, then filtering compares the template and query distributions over the $k$ features. We show that state-of-the-art performance was achieved in low-resolution data, and our method outperforms previous methods at higher resolution showing the robustness and scalability of the approach.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023



Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Interpreting deep learning output for out-of-distribution detection
Nov 07, 2022Abstract:Commonly used AI networks are very self-confident in their predictions, even when the evidence for a certain decision is dubious. The investigation of a deep learning model output is pivotal for understanding its decision processes and assessing its capabilities and limitations. By analyzing the distributions of raw network output vectors, it can be observed that each class has its own decision boundary and, thus, the same raw output value has different support for different classes. Inspired by this fact, we have developed a new method for out-of-distribution detection. The method offers an explanatory step beyond simple thresholding of the softmax output towards understanding and interpretation of the model learning process and its output. Instead of assigning the class label of the highest logit to each new sample presented to the network, it takes the distributions over all classes into consideration. A probability score interpreter (PSI) is created based on the joint logit values in relation to their respective correct vs wrong class distributions. The PSI suggests whether the sample is likely to belong to a specific class, whether the network is unsure, or whether the sample is likely an outlier or unknown type for the network. The simple PSI has the benefit of being applicable on already trained networks. The distributions for correct vs wrong class for each output node are established by simply running the training examples through the trained network. We demonstrate our OOD detection method on a challenging transmission electron microscopy virus image dataset. We simulate a real-world application in which images of virus types unknown to a trained virus classifier, yet acquired with the same procedures and instruments, constitute the OOD samples.
Towards Better Guided Attention and Human Knowledge Insertion in Deep Convolutional Neural Networks
Oct 20, 2022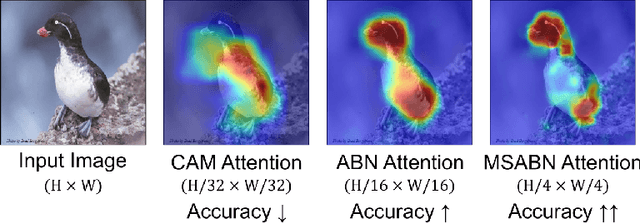

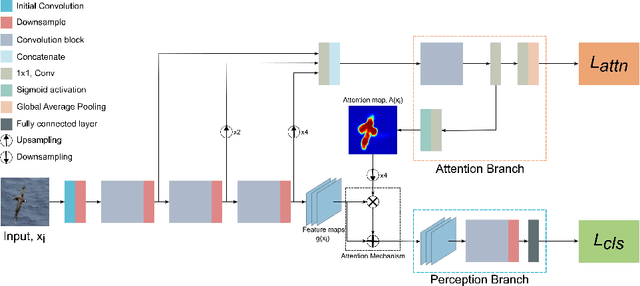
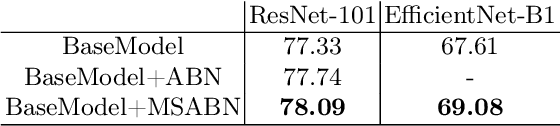
Abstract:Attention Branch Networks (ABNs) have been shown to simultaneously provide visual explanation and improve the performance of deep convolutional neural networks (CNNs). In this work, we introduce Multi-Scale Attention Branch Networks (MSABN), which enhance the resolution of the generated attention maps, and improve the performance. We evaluate MSABN on benchmark image recognition and fine-grained recognition datasets where we observe MSABN outperforms ABN and baseline models. We also introduce a new data augmentation strategy utilizing the attention maps to incorporate human knowledge in the form of bounding box annotations of the objects of interest. We show that even with a limited number of edited samples, a significant performance gain can be achieved with this strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge