Hui Qiao
SegEarth-R2: Towards Comprehensive Language-guided Segmentation for Remote Sensing Images
Dec 23, 2025Abstract:Effectively grounding complex language to pixels in remote sensing (RS) images is a critical challenge for applications like disaster response and environmental monitoring. Current models can parse simple, single-target commands but fail when presented with complex geospatial scenarios, e.g., segmenting objects at various granularities, executing multi-target instructions, and interpreting implicit user intent. To drive progress against these failures, we present LaSeRS, the first large-scale dataset built for comprehensive training and evaluation across four critical dimensions of language-guided segmentation: hierarchical granularity, target multiplicity, reasoning requirements, and linguistic variability. By capturing these dimensions, LaSeRS moves beyond simple commands, providing a benchmark for complex geospatial reasoning. This addresses a critical gap: existing datasets oversimplify, leading to sensitivity-prone real-world models. We also propose SegEarth-R2, an MLLM architecture designed for comprehensive language-guided segmentation in RS, which directly confronts these challenges. The model's effectiveness stems from two key improvements: (1) a spatial attention supervision mechanism specifically handles the localization of small objects and their components, and (2) a flexible and efficient segmentation query mechanism that handles both single-target and multi-target scenarios. Experimental results demonstrate that our SegEarth-R2 achieves outstanding performance on LaSeRS and other benchmarks, establishing a powerful baseline for the next generation of geospatial segmentation. All data and code will be released at https://github.com/earth-insights/SegEarth-R2.
ZoomEarth: Active Perception for Ultra-High-Resolution Geospatial Vision-Language Tasks
Nov 15, 2025



Abstract:Ultra-high-resolution (UHR) remote sensing (RS) images offer rich fine-grained information but also present challenges in effective processing. Existing dynamic resolution and token pruning methods are constrained by a passive perception paradigm, suffering from increased redundancy when obtaining finer visual inputs. In this work, we explore a new active perception paradigm that enables models to revisit information-rich regions. First, we present LRS-GRO, a large-scale benchmark dataset tailored for active perception in UHR RS processing, encompassing 17 question types across global, region, and object levels, annotated via a semi-automatic pipeline. Building on LRS-GRO, we propose ZoomEarth, an adaptive cropping-zooming framework with a novel Region-Guided reward that provides fine-grained guidance. Trained via supervised fine-tuning (SFT) and Group Relative Policy Optimization (GRPO), ZoomEarth achieves state-of-the-art performance on LRS-GRO and, in the zero-shot setting, on three public UHR remote sensing benchmarks. Furthermore, ZoomEarth can be seamlessly integrated with downstream models for tasks such as cloud removal, denoising, segmentation, and image editing through simple tool interfaces, demonstrating strong versatility and extensibility.
V2V3D: View-to-View Denoised 3D Reconstruction for Light-Field Microscopy
Apr 10, 2025



Abstract:Light field microscopy (LFM) has gained significant attention due to its ability to capture snapshot-based, large-scale 3D fluorescence images. However, existing LFM reconstruction algorithms are highly sensitive to sensor noise or require hard-to-get ground-truth annotated data for training. To address these challenges, this paper introduces V2V3D, an unsupervised view2view-based framework that establishes a new paradigm for joint optimization of image denoising and 3D reconstruction in a unified architecture. We assume that the LF images are derived from a consistent 3D signal, with the noise in each view being independent. This enables V2V3D to incorporate the principle of noise2noise for effective denoising. To enhance the recovery of high-frequency details, we propose a novel wave-optics-based feature alignment technique, which transforms the point spread function, used for forward propagation in wave optics, into convolution kernels specifically designed for feature alignment. Moreover, we introduce an LFM dataset containing LF images and their corresponding 3D intensity volumes. Extensive experiments demonstrate that our approach achieves high computational efficiency and outperforms the other state-of-the-art methods. These advancements position V2V3D as a promising solution for 3D imaging under challenging conditions.
PNR: Physics-informed Neural Representation for high-resolution LFM reconstruction
Sep 26, 2024Abstract:Light field microscopy (LFM) has been widely utilized in various fields for its capability to efficiently capture high-resolution 3D scenes. Despite the rapid advancements in neural representations, there are few methods specifically tailored for microscopic scenes. Existing approaches often do not adequately address issues such as the loss of high-frequency information due to defocus and sample aberration, resulting in suboptimal performance. In addition, existing methods, including RLD, INR, and supervised U-Net, face challenges such as sensitivity to initial estimates, reliance on extensive labeled data, and low computational efficiency, all of which significantly diminish the practicality in complex biological scenarios. This paper introduces PNR (Physics-informed Neural Representation), a method for high-resolution LFM reconstruction that significantly enhances performance. Our method incorporates an unsupervised and explicit feature representation approach, resulting in a 6.1 dB improvement in PSNR than RLD. Additionally, our method employs a frequency-based training loss, enabling better recovery of high-frequency details, which leads to a reduction in LPIPS by at least half compared to SOTA methods (1.762 V.S. 3.646 of DINER). Moreover, PNR integrates a physics-informed aberration correction strategy that optimizes Zernike polynomial parameters during optimization, thereby reducing the information loss caused by aberrations and improving spatial resolution. These advancements make PNR a promising solution for long-term high-resolution biological imaging applications. Our code and dataset will be made publicly available.
Prostate cancer histopathology with label-free multispectral deep UV microscopy quantifies phenotypes of tumor grade and aggressiveness
Jun 01, 2021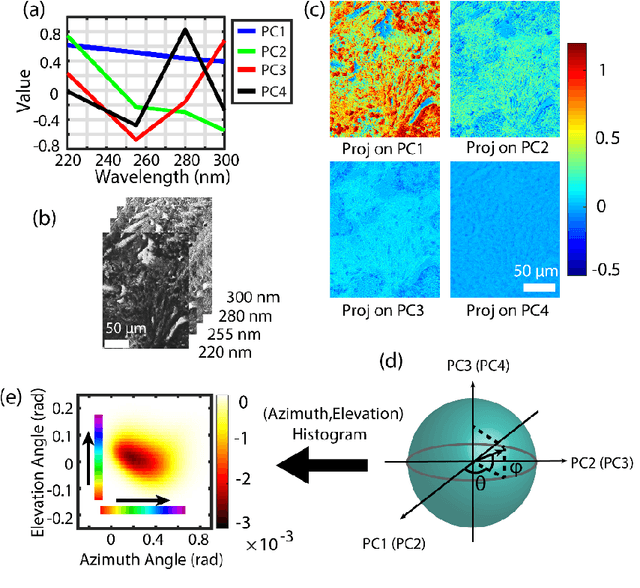
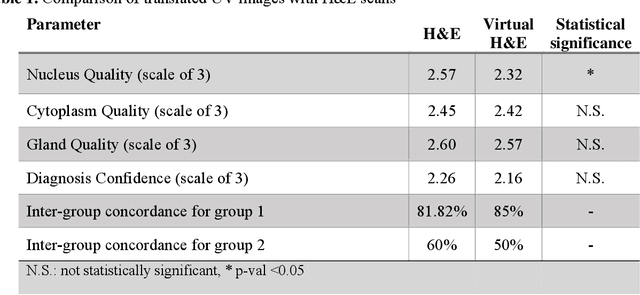


Abstract:Identifying prostate cancer patients that are harboring aggressive forms of prostate cancer remains a significant clinical challenge. To shed light on this problem, we develop an approach based on multispectral deep-ultraviolet (UV) microscopy that provides novel quantitative insight into the aggressiveness and grade of this disease. First, we find that UV spectral signatures from endogenous molecules give rise to a phenotypical continuum that differentiates critical structures of thin tissue sections with subcellular spatial resolution, including nuclei, cytoplasm, stroma, basal cells, nerves, and inflammation. Further, we show that this phenotypical continuum can be applied as a surrogate biomarker of prostate cancer malignancy, where patients with the most aggressive tumors show a ubiquitous glandular phenotypical shift. Lastly, we adapt a two-part Cycle-consistent Generative Adversarial Network to translate the label-free deep-UV images into virtual hematoxylin and eosin (H&E) stained images. Agreement between the virtual H&E images and the gold standard H&E-stained tissue sections is evaluated by a panel of pathologists who find that the two modalities are in excellent agreement. This work has significant implications towards improving our ability to objectively quantify prostate cancer grade and aggressiveness, thus improving the management and clinical outcomes of prostate cancer patients. This same approach can also be applied broadly in other tumor types to achieve low-cost, stain-free, quantitative histopathological analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge