Hongyu Yuan
MAGE-KT: Multi-Agent Graph-Enhanced Knowledge Tracing with Subgraph Retrieval and Asymmetric Fusion
Jan 23, 2026Abstract:Knowledge Tracing (KT) aims to model a student's learning trajectory and predict performance on the next question. A key challenge is how to better represent the relationships among students, questions, and knowledge concepts (KCs). Recently, graph-based KT paradigms have shown promise for this problem. However, existing methods have not sufficiently explored inter-concept relations, often inferred solely from interaction sequences. In addition, the scale and heterogeneity of KT graphs make full-graph encoding both computationally both costly and noise-prone, causing attention to bleed into student-irrelevant regions and degrading the fidelity of inter-KC relations. To address these issues, we propose a novel framework: Multi-Agent Graph-Enhanced Knowledge Tracing (MAGE-KT). It constructs a multi-view heterogeneous graph by combining a multi-agent KC relation extractor and a student-question interaction graph, capturing complementary semantic and behavioral signals. Conditioned on the target student's history, it retrieves compact, high-value subgraphs and integrates them using an Asymmetric Cross-attention Fusion Module to enhance prediction while avoiding attention diffusion and irrelevant computation. Experiments on three widely used KT datasets show substantial improvements in KC-relation accuracy and clear gains in next-question prediction over existing methods.
Advancing Multimodal Teacher Sentiment Analysis:The Large-Scale T-MED Dataset & The Effective AAM-TSA Model
Dec 23, 2025



Abstract:Teachers' emotional states are critical in educational scenarios, profoundly impacting teaching efficacy, student engagement, and learning achievements. However, existing studies often fail to accurately capture teachers' emotions due to the performative nature and overlook the critical impact of instructional information on emotional expression.In this paper, we systematically investigate teacher sentiment analysis by building both the dataset and the model accordingly. We construct the first large-scale teacher multimodal sentiment analysis dataset, T-MED.To ensure labeling accuracy and efficiency, we employ a human-machine collaborative labeling process.The T-MED dataset includes 14,938 instances of teacher emotional data from 250 real classrooms across 11 subjects ranging from K-12 to higher education, integrating multimodal text, audio, video, and instructional information.Furthermore, we propose a novel asymmetric attention-based multimodal teacher sentiment analysis model, AAM-TSA.AAM-TSA introduces an asymmetric attention mechanism and hierarchical gating unit to enable differentiated cross-modal feature fusion and precise emotional classification. Experimental results demonstrate that AAM-TSA significantly outperforms existing state-of-the-art methods in terms of accuracy and interpretability on the T-MED dataset.
HISE-KT: Synergizing Heterogeneous Information Networks and LLMs for Explainable Knowledge Tracing with Meta-Path Optimization
Nov 19, 2025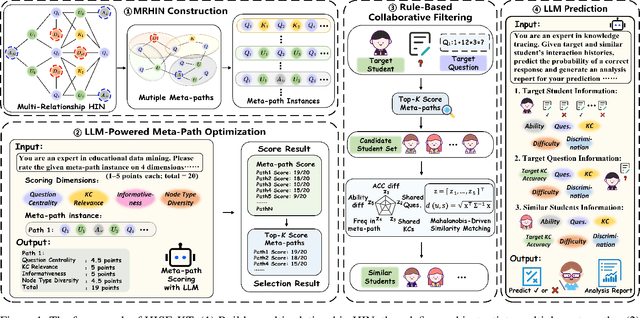
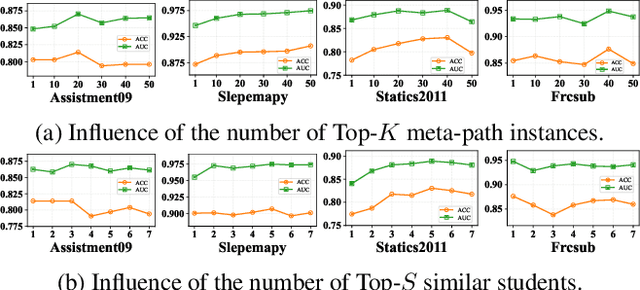
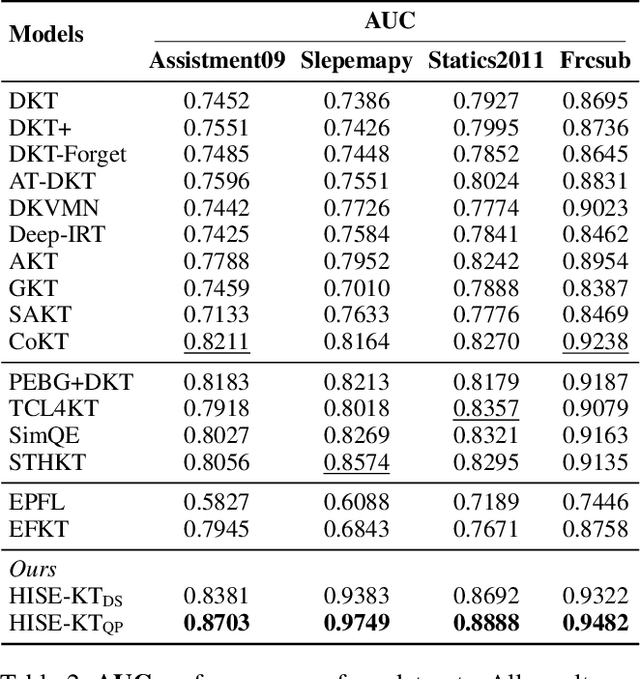
Abstract:Knowledge Tracing (KT) aims to mine students' evolving knowledge states and predict their future question-answering performance. Existing methods based on heterogeneous information networks (HINs) are prone to introducing noises due to manual or random selection of meta-paths and lack necessary quality assessment of meta-path instances. Conversely, recent large language models (LLMs)-based methods ignore the rich information across students, and both paradigms struggle to deliver consistently accurate and evidence-based explanations. To address these issues, we propose an innovative framework, HIN-LLM Synergistic Enhanced Knowledge Tracing (HISE-KT), which seamlessly integrates HINs with LLMs. HISE-KT first builds a multi-relationship HIN containing diverse node types to capture the structural relations through multiple meta-paths. The LLM is then employed to intelligently score and filter meta-path instances and retain high-quality paths, pioneering automated meta-path quality assessment. Inspired by educational psychology principles, a similar student retrieval mechanism based on meta-paths is designed to provide a more valuable context for prediction. Finally, HISE-KT uses a structured prompt to integrate the target student's history with the retrieved similar trajectories, enabling the LLM to generate not only accurate predictions but also evidence-backed, explainable analysis reports. Experiments on four public datasets show that HISE-KT outperforms existing KT baselines in both prediction performance and interpretability.
Nonhuman Primate Brain Tissue Segmentation Using a Transfer Learning Approach
Apr 01, 2025Abstract:Non-human primates (NHPs) serve as critical models for understanding human brain function and neurological disorders due to their close evolutionary relationship with humans. Accurate brain tissue segmentation in NHPs is critical for understanding neurological disorders, but challenging due to the scarcity of annotated NHP brain MRI datasets, the small size of the NHP brain, the limited resolution of available imaging data and the anatomical differences between human and NHP brains. To address these challenges, we propose a novel approach utilizing STU-Net with transfer learning to leverage knowledge transferred from human brain MRI data to enhance segmentation accuracy in the NHP brain MRI, particularly when training data is limited. The combination of STU-Net and transfer learning effectively delineates complex tissue boundaries and captures fine anatomical details specific to NHP brains. Notably, our method demonstrated improvement in segmenting small subcortical structures such as putamen and thalamus that are challenging to resolve with limited spatial resolution and tissue contrast, and achieved DSC of over 0.88, IoU over 0.8 and HD95 under 7. This study introduces a robust method for multi-class brain tissue segmentation in NHPs, potentially accelerating research in evolutionary neuroscience and preclinical studies of neurological disorders relevant to human health.
Listening and Seeing Again: Generative Error Correction for Audio-Visual Speech Recognition
Jan 03, 2025



Abstract:Unlike traditional Automatic Speech Recognition (ASR), Audio-Visual Speech Recognition (AVSR) takes audio and visual signals simultaneously to infer the transcription. Recent studies have shown that Large Language Models (LLMs) can be effectively used for Generative Error Correction (GER) in ASR by predicting the best transcription from ASR-generated N-best hypotheses. However, these LLMs lack the ability to simultaneously understand audio and visual, making the GER approach challenging to apply in AVSR. In this work, we propose a novel GER paradigm for AVSR, termed AVGER, that follows the concept of ``listening and seeing again''. Specifically, we first use the powerful AVSR system to read the audio and visual signals to get the N-Best hypotheses, and then use the Q-former-based Multimodal Synchronous Encoder to read the audio and visual information again and convert them into an audio and video compression representation respectively that can be understood by LLM. Afterward, the audio-visual compression representation and the N-Best hypothesis together constitute a Cross-modal Prompt to guide the LLM in producing the best transcription. In addition, we also proposed a Multi-Level Consistency Constraint training criterion, including logits-level, utterance-level and representations-level, to improve the correction accuracy while enhancing the interpretability of audio and visual compression representations. The experimental results on the LRS3 dataset show that our method outperforms current mainstream AVSR systems. The proposed AVGER can reduce the Word Error Rate (WER) by 24% compared to them. Code and models can be found at: https://github.com/CircleRedRain/AVGER.
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023



Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge