Georges El Fakhri
Generative Self-training for Cross-domain Unsupervised Tagged-to-Cine MRI Synthesis
Jun 23, 2021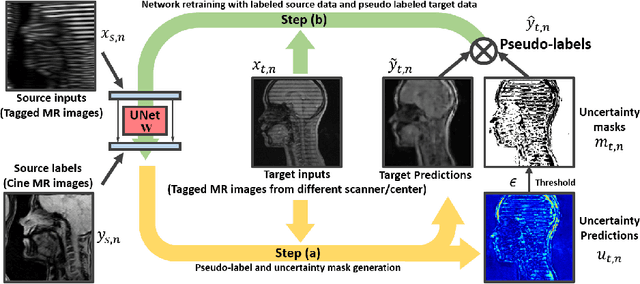
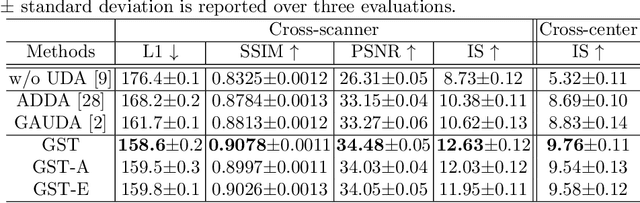
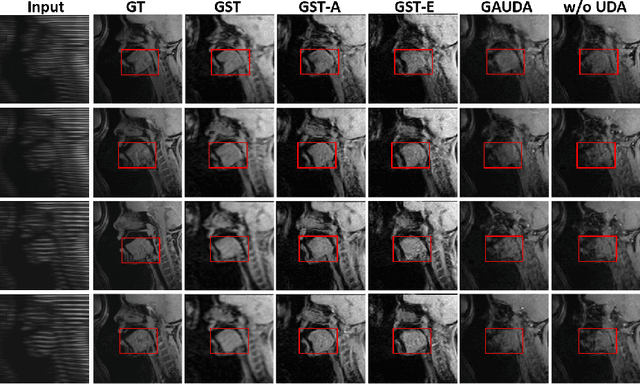
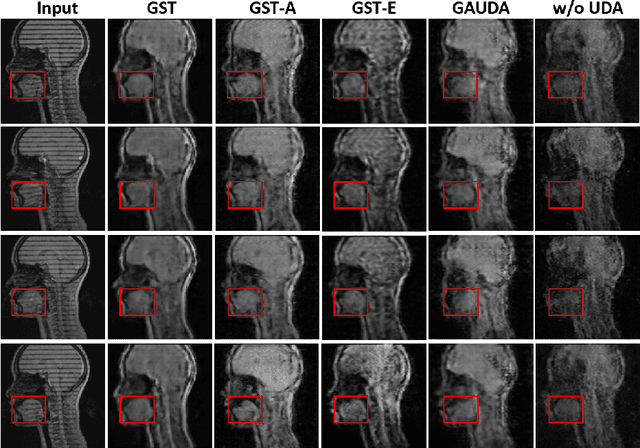
Abstract:Self-training based unsupervised domain adaptation (UDA) has shown great potential to address the problem of domain shift, when applying a trained deep learning model in a source domain to unlabeled target domains. However, while the self-training UDA has demonstrated its effectiveness on discriminative tasks, such as classification and segmentation, via the reliable pseudo-label selection based on the softmax discrete histogram, the self-training UDA for generative tasks, such as image synthesis, is not fully investigated. In this work, we propose a novel generative self-training (GST) UDA framework with continuous value prediction and regression objective for cross-domain image synthesis. Specifically, we propose to filter the pseudo-label with an uncertainty mask, and quantify the predictive confidence of generated images with practical variational Bayes learning. The fast test-time adaptation is achieved by a round-based alternative optimization scheme. We validated our framework on the tagged-to-cine magnetic resonance imaging (MRI) synthesis problem, where datasets in the source and target domains were acquired from different scanners or centers. Extensive validations were carried out to verify our framework against popular adversarial training UDA methods. Results show that our GST, with tagged MRI of test subjects in new target domains, improved the synthesis quality by a large margin, compared with the adversarial training UDA methods.
Adapting Off-the-Shelf Source Segmenter for Target Medical Image Segmentation
Jun 23, 2021
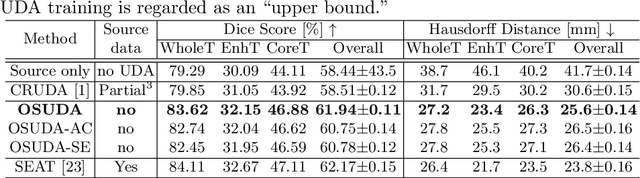
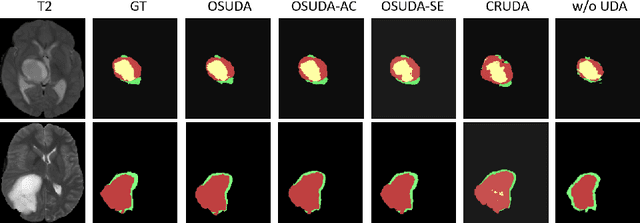
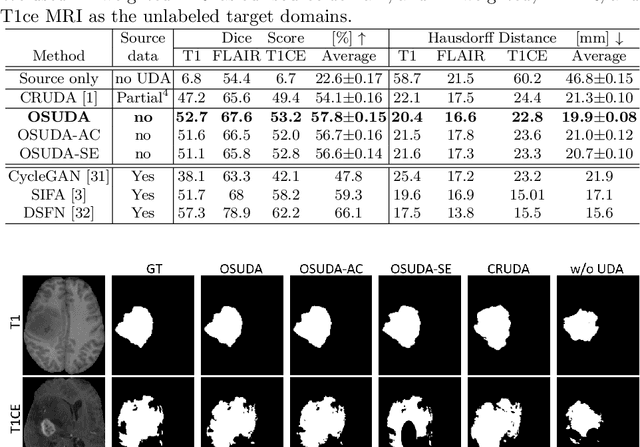
Abstract:Unsupervised domain adaptation (UDA) aims to transfer knowledge learned from a labeled source domain to an unlabeled and unseen target domain, which is usually trained on data from both domains. Access to the source domain data at the adaptation stage, however, is often limited, due to data storage or privacy issues. To alleviate this, in this work, we target source free UDA for segmentation, and propose to adapt an ``off-the-shelf" segmentation model pre-trained in the source domain to the target domain, with an adaptive batch-wise normalization statistics adaptation framework. Specifically, the domain-specific low-order batch statistics, i.e., mean and variance, are gradually adapted with an exponential momentum decay scheme, while the consistency of domain shareable high-order batch statistics, i.e., scaling and shifting parameters, is explicitly enforced by our optimization objective. The transferability of each channel is adaptively measured first from which to balance the contribution of each channel. Moreover, the proposed source free UDA framework is orthogonal to unsupervised learning methods, e.g., self-entropy minimization, which can thus be simply added on top of our framework. Extensive experiments on the BraTS 2018 database show that our source free UDA framework outperformed existing source-relaxed UDA methods for the cross-subtype UDA segmentation task and yielded comparable results for the cross-modality UDA segmentation task, compared with a supervised UDA methods with the source data.
Dual-cycle Constrained Bijective VAE-GAN For Tagged-to-Cine Magnetic Resonance Image Synthesis
Jan 14, 2021
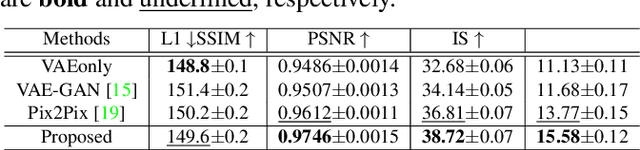

Abstract:Tagged magnetic resonance imaging (MRI) is a widely used imaging technique for measuring tissue deformation in moving organs. Due to tagged MRI's intrinsic low anatomical resolution, another matching set of cine MRI with higher resolution is sometimes acquired in the same scanning session to facilitate tissue segmentation, thus adding extra time and cost. To mitigate this, in this work, we propose a novel dual-cycle constrained bijective VAE-GAN approach to carry out tagged-to-cine MR image synthesis. Our method is based on a variational autoencoder backbone with cycle reconstruction constrained adversarial training to yield accurate and realistic cine MR images given tagged MR images. Our framework has been trained, validated, and tested using 1,768, 416, and 1,560 subject-independent paired slices of tagged and cine MRI from twenty healthy subjects, respectively, demonstrating superior performance over the comparison methods. Our method can potentially be used to reduce the extra acquisition time and cost, while maintaining the same workflow for further motion analyses.
A Unified Conditional Disentanglement Framework for Multimodal Brain MR Image Translation
Jan 14, 2021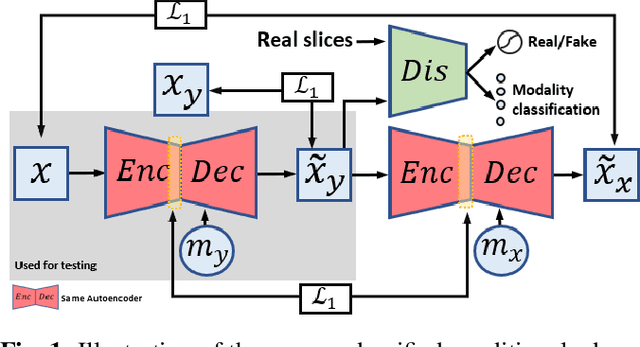
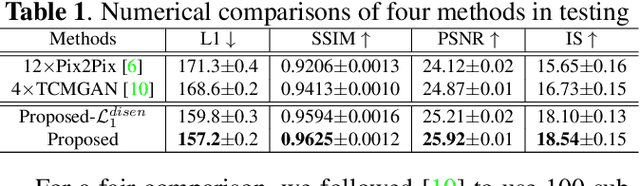
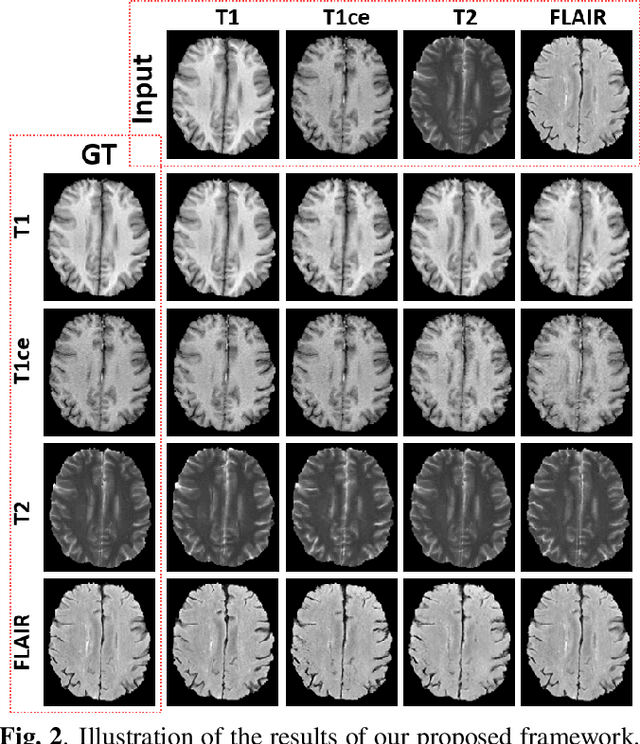
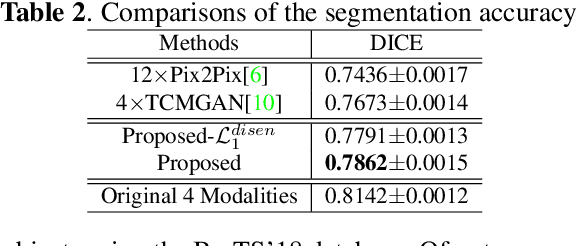
Abstract:Multimodal MRI provides complementary and clinically relevant information to probe tissue condition and to characterize various diseases. However, it is often difficult to acquire sufficiently many modalities from the same subject due to limitations in study plans, while quantitative analysis is still demanded. In this work, we propose a unified conditional disentanglement framework to synthesize any arbitrary modality from an input modality. Our framework hinges on a cycle-constrained conditional adversarial training approach, where it can extract a modality-invariant anatomical feature with a modality-agnostic encoder and generate a target modality with a conditioned decoder. We validate our framework on four MRI modalities, including T1-weighted, T1 contrast enhanced, T2-weighted, and FLAIR MRI, from the BraTS'18 database, showing superior performance on synthesis quality over the comparison methods. In addition, we report results from experiments on a tumor segmentation task carried out with synthesized data.
VoxelHop: Successive Subspace Learning for ALS Disease Classification Using Structural MRI
Jan 13, 2021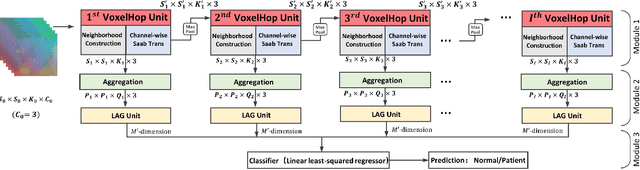
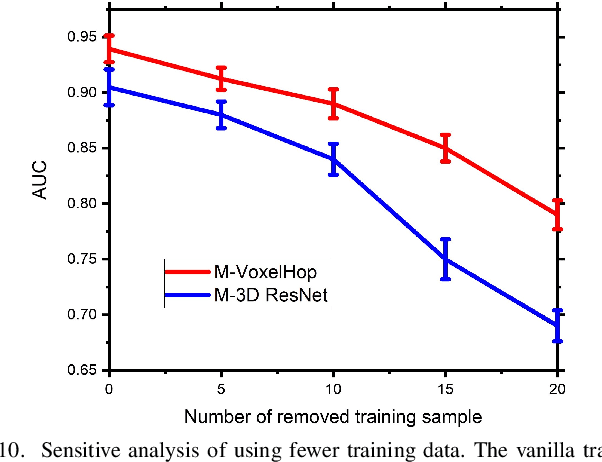
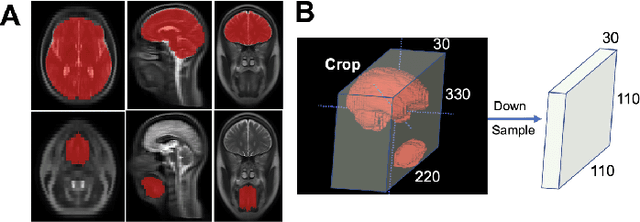
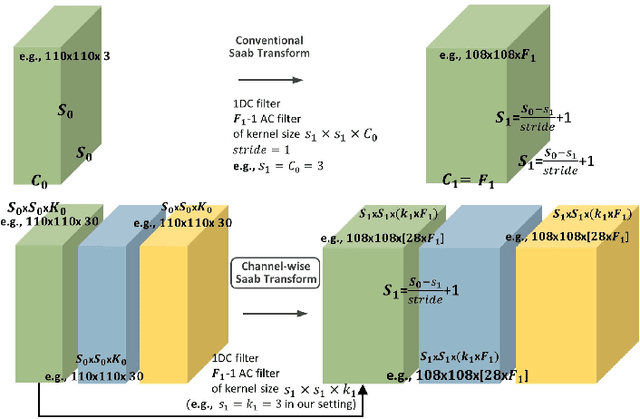
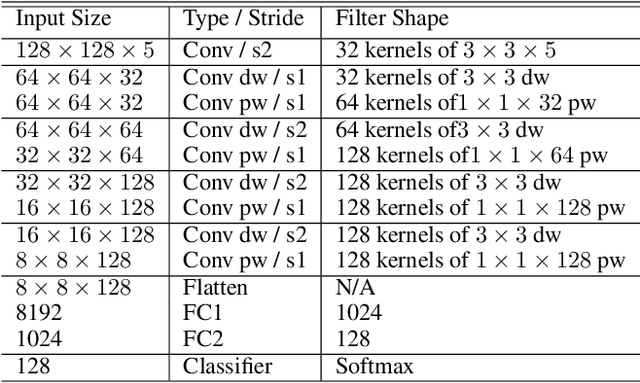
Abstract:Deep learning has great potential for accurate detection and classification of diseases with medical imaging data, but the performance is often limited by the number of training datasets and memory requirements. In addition, many deep learning models are considered a "black-box," thereby often limiting their adoption in clinical applications. To address this, we present a successive subspace learning model, termed VoxelHop, for accurate classification of Amyotrophic Lateral Sclerosis (ALS) using T2-weighted structural MRI data. Compared with popular convolutional neural network (CNN) architectures, VoxelHop has modular and transparent structures with fewer parameters without any backpropagation, so it is well-suited to small dataset size and 3D imaging data. Our VoxelHop has four key components, including (1) sequential expansion of near-to-far neighborhood for multi-channel 3D data; (2) subspace approximation for unsupervised dimension reduction; (3) label-assisted regression for supervised dimension reduction; and (4) concatenation of features and classification between controls and patients. Our experimental results demonstrate that our framework using a total of 20 controls and 26 patients achieves an accuracy of 93.48$\%$ and an AUC score of 0.9394 in differentiating patients from controls, even with a relatively small number of datasets, showing its robustness and effectiveness. Our thorough evaluations also show its validity and superiority to the state-of-the-art 3D CNN classification methods. Our framework can easily be generalized to other classification tasks using different imaging modalities.
Subtype-aware Unsupervised Domain Adaptation for Medical Diagnosis
Jan 11, 2021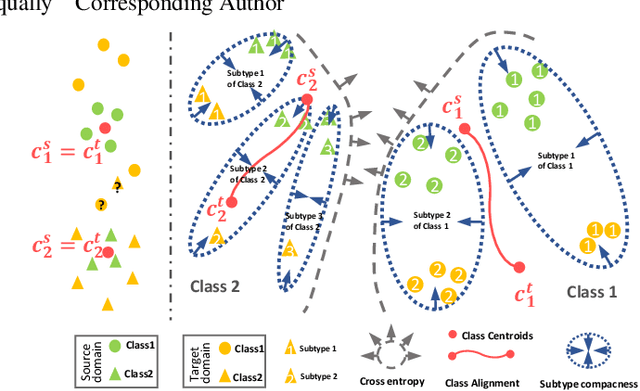
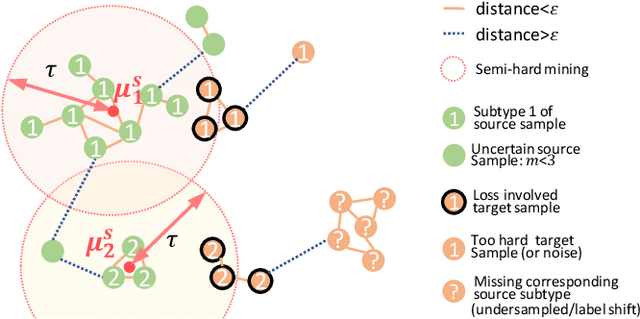
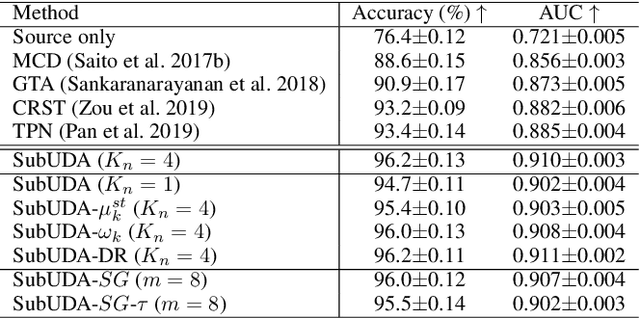
Abstract:Recent advances in unsupervised domain adaptation (UDA) show that transferable prototypical learning presents a powerful means for class conditional alignment, which encourages the closeness of cross-domain class centroids. However, the cross-domain inner-class compactness and the underlying fine-grained subtype structure remained largely underexplored. In this work, we propose to adaptively carry out the fine-grained subtype-aware alignment by explicitly enforcing the class-wise separation and subtype-wise compactness with intermediate pseudo labels. Our key insight is that the unlabeled subtypes of a class can be divergent to one another with different conditional and label shifts, while inheriting the local proximity within a subtype. The cases of with or without the prior information on subtype numbers are investigated to discover the underlying subtype structure in an online fashion. The proposed subtype-aware dynamic UDA achieves promising results on medical diagnosis tasks.
A Deep Joint Sparse Non-negative Matrix Factorization Framework for Identifying the Common and Subject-specific Functional Units of Tongue Motion During Speech
Jul 09, 2020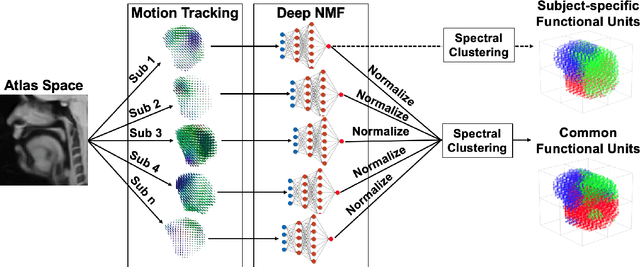
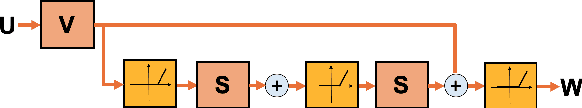
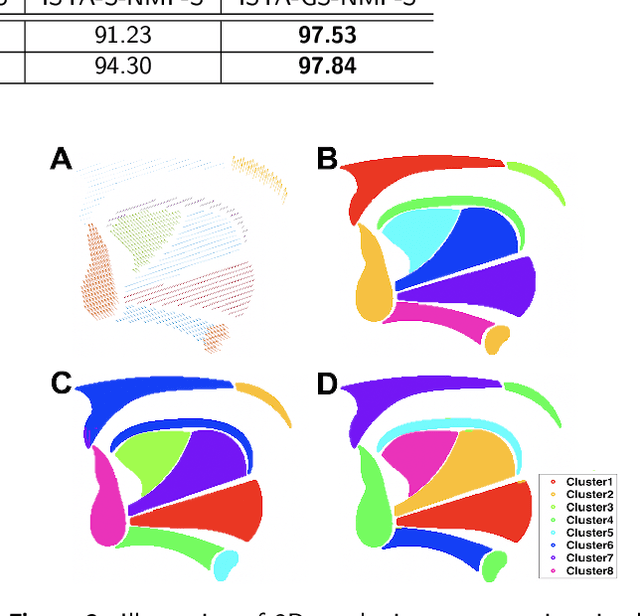
Abstract:Intelligible speech is produced by creating varying internal local muscle groupings---i.e., functional units---that are generated in a systematic and coordinated manner. There are two major challenges in characterizing and analyzing functional units. First, due to the complex and convoluted nature of tongue structure and function, it is of great importance to develop a method that can accurately decode complex muscle coordination patterns during speech. Second, it is challenging to keep identified functional units across subjects comparable due to their substantial variability. In this work, to address these challenges, we develop a new deep learning framework to identify common and subject-specific functional units of tongue motion during speech. Our framework hinges on joint deep graph-regularized sparse non-negative matrix factorization (NMF) using motion quantities derived from displacements by tagged Magnetic Resonance Imaging. More specifically, we transform NMF with sparse and manifold regularizations into modular architectures akin to deep neural networks by means of unfolding the Iterative Shrinkage-Thresholding Algorithm to learn interpretable building blocks and associated weighting map. We then apply spectral clustering to common and subject-specific functional units. Experiments carried out with simulated datasets show that the proposed method surpasses the comparison methods. Experiments carried out with in vivo tongue motion datasets show that the proposed method can determine the common and subject-specific functional units with increased interpretability and decreased size variability.
End-to-end Lung Nodule Detection in Computed Tomography
Oct 02, 2018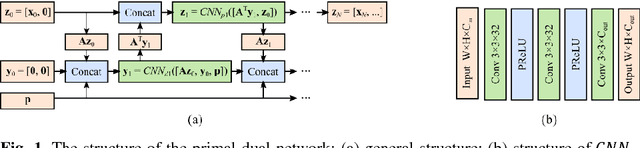
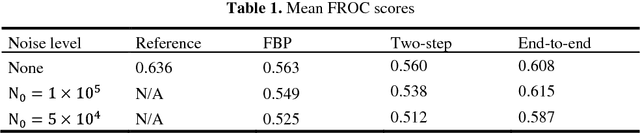
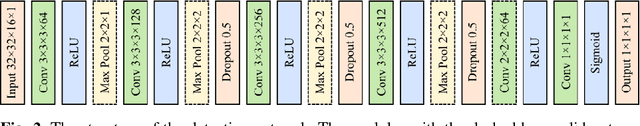
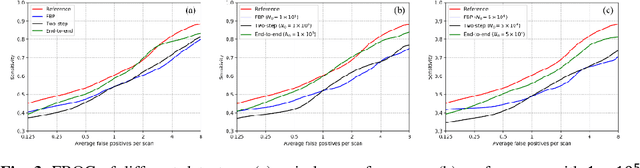
Abstract:Computer aided diagnostic (CAD) system is crucial for modern med-ical imaging. But almost all CAD systems operate on reconstructed images, which were optimized for radiologists. Computer vision can capture features that is subtle to human observers, so it is desirable to design a CAD system op-erating on the raw data. In this paper, we proposed a deep-neural-network-based detection system for lung nodule detection in computed tomography (CT). A primal-dual-type deep reconstruction network was applied first to convert the raw data to the image space, followed by a 3-dimensional convolutional neural network (3D-CNN) for the nodule detection. For efficient network training, the deep reconstruction network and the CNN detector was trained sequentially first, then followed by one epoch of end-to-end fine tuning. The method was evaluated on the Lung Image Database Consortium image collection (LIDC-IDRI) with simulated forward projections. With 144 multi-slice fanbeam pro-jections, the proposed end-to-end detector could achieve comparable sensitivity with the reference detector, which was trained and applied on the fully-sampled image data. It also demonstrated superior detection performance compared to detectors trained on the reconstructed images. The proposed method is general and could be expanded to most detection tasks in medical imaging.
A Sparse Non-negative Matrix Factorization Framework for Identifying Functional Units of Tongue Behavior from MRI
Sep 29, 2018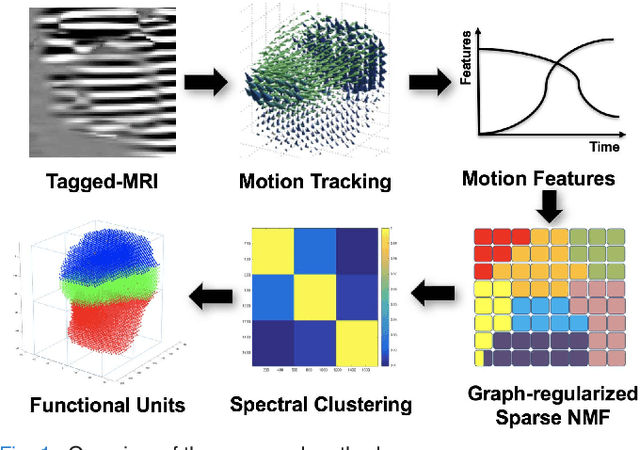
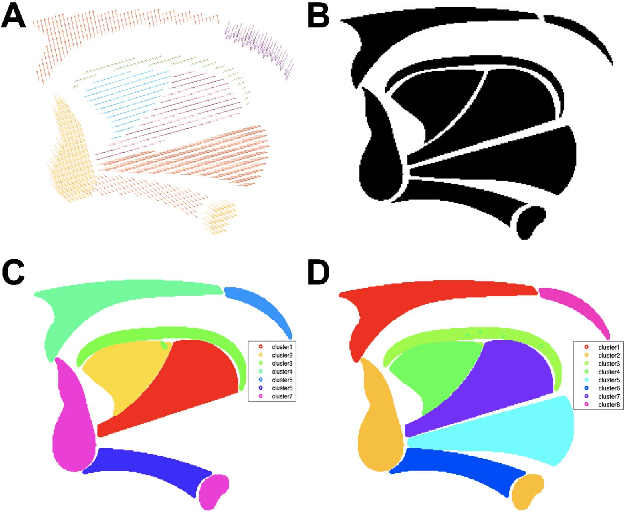
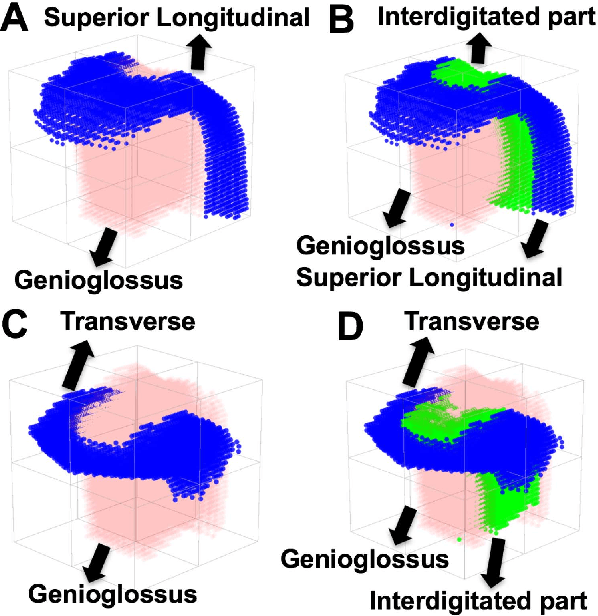
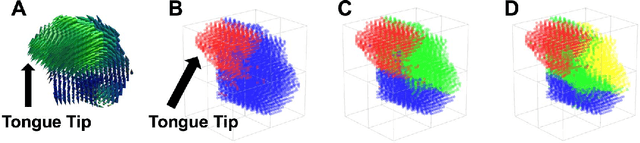
Abstract:Muscle coordination patterns of lingual behaviors are synergies generated by deforming local muscle groups in a variety of ways. Functional units are functional muscle groups of local structural elements within the tongue that compress, expand, and move in a cohesive and consistent manner. Identifying the functional units using tagged-Magnetic Resonance Imaging (MRI) sheds light on the mechanisms of normal and pathological muscle coordination patterns, yielding improvement in surgical planning, treatment, or rehabilitation procedures. Here, to mine this information, we propose a matrix factorization and probabilistic graphical model framework to produce building blocks and their associated weighting map using motion quantities extracted from tagged-MRI. Our tagged-MRI imaging and accurate voxel-level tracking provide previously unavailable internal tongue motion patterns, thus revealing the inner workings of the tongue during speech or other lingual behaviors. We then employ spectral clustering on the weighting map to identify the cohesive regions defined by the tongue motion that may involve multiple or undocumented regions. To evaluate our method, we perform a series of experiments. We first use two-dimensional images and synthetic data to demonstrate the accuracy of our method. We then use three-dimensional synthetic and \textit{in vivo} tongue motion data using protrusion and simple speech tasks to identify subject-specific and data-driven functional units of the tongue in localized regions.
Speech Map: A Statistical Multimodal Atlas of 4D Tongue Motion During Speech from Tagged and Cine MR Images
Sep 15, 2018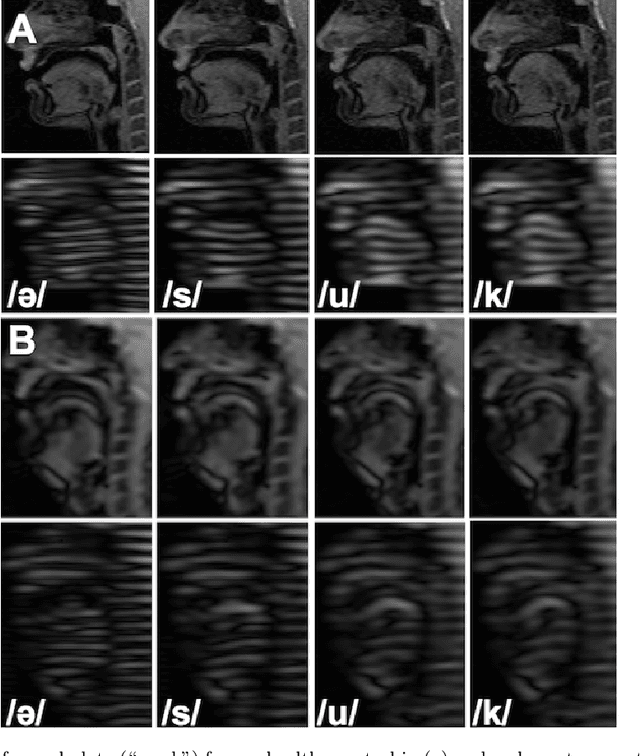

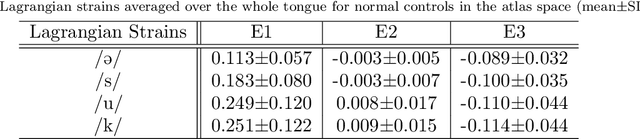
Abstract:Quantitative measurement of functional and anatomical traits of 4D tongue motion in the course of speech or other lingual behaviors remains a major challenge in scientific research and clinical applications. Here, we introduce a statistical multimodal atlas of 4D tongue motion using healthy subjects, which enables a combined quantitative characterization of tongue motion in a reference anatomical configuration. This atlas framework, termed Speech Map, combines cine- and tagged-MRI in order to provide both the anatomic reference and motion information during speech. Our approach involves a series of steps including (1) construction of a common reference anatomical configuration from cine-MRI, (2) motion estimation from tagged-MRI, (3) transformation of the motion estimations to the reference anatomical configuration, and (4) computation of motion quantities such as Lagrangian strain. Using this framework, the anatomic configuration of the tongue appears motionless, while the motion fields and associated strain measurements change over the time course of speech. In addition, to form a succinct representation of the high-dimensional and complex motion fields, principal component analysis is carried out to characterize the central tendencies and variations of motion fields of our speech tasks. Our proposed method provides a platform to quantitatively and objectively explain the differences and variability of tongue motion by illuminating internal motion and strain that have so far been intractable. The findings are used to understand how tongue function for speech is limited by abnormal internal motion and strain in glossectomy patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge