Eddie Calleja
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024



Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
DeepRacer: Educational Autonomous Racing Platform for Experimentation with Sim2Real Reinforcement Learning
Nov 05, 2019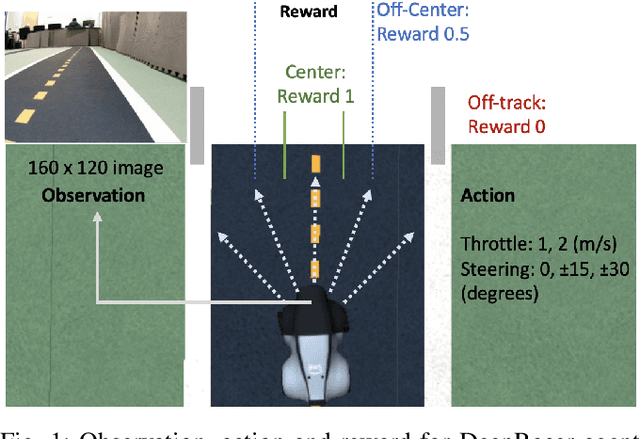
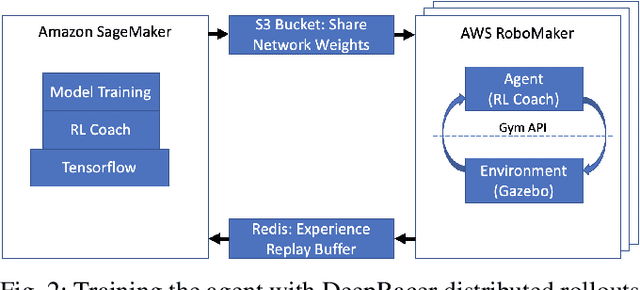
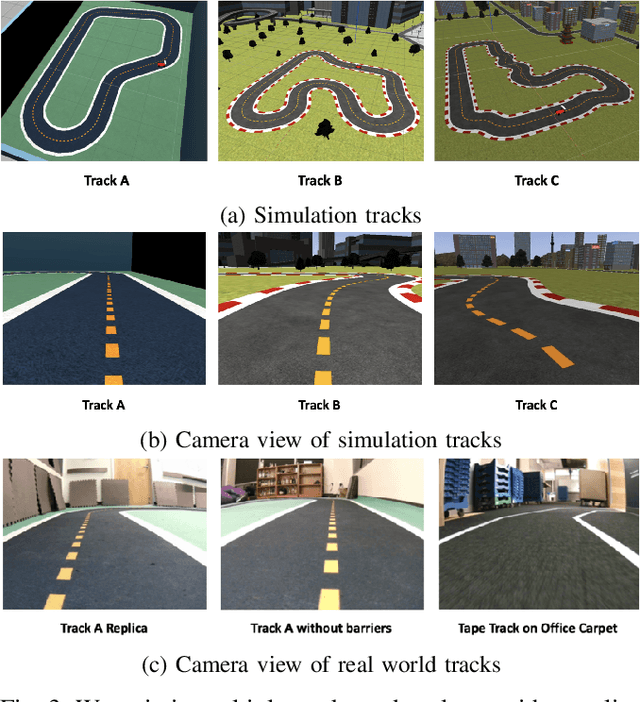

Abstract:DeepRacer is a platform for end-to-end experimentation with RL and can be used to systematically investigate the key challenges in developing intelligent control systems. Using the platform, we demonstrate how a 1/18th scale car can learn to drive autonomously using RL with a monocular camera. It is trained in simulation with no additional tuning in physical world and demonstrates: 1) formulation and solution of a robust reinforcement learning algorithm, 2) narrowing the reality gap through joint perception and dynamics, 3) distributed on-demand compute architecture for training optimal policies, and 4) a robust evaluation method to identify when to stop training. It is the first successful large-scale deployment of deep reinforcement learning on a robotic control agent that uses only raw camera images as observations and a model-free learning method to perform robust path planning. We open source our code and video demo on GitHub: https://git.io/fjxoJ.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge