Dina Demner-Fushman
A Dataset and Benchmark for Consumer Healthcare Question Summarization
Dec 29, 2025Abstract:The quest for seeking health information has swamped the web with consumers health-related questions. Generally, consumers use overly descriptive and peripheral information to express their medical condition or other healthcare needs, contributing to the challenges of natural language understanding. One way to address this challenge is to summarize the questions and distill the key information of the original question. Recently, large-scale datasets have significantly propelled the development of several summarization tasks, such as multi-document summarization and dialogue summarization. However, a lack of a domain-expert annotated dataset for the consumer healthcare questions summarization task inhibits the development of an efficient summarization system. To address this issue, we introduce a new dataset, CHQ-Sum,m that contains 1507 domain-expert annotated consumer health questions and corresponding summaries. The dataset is derived from the community question answering forum and therefore provides a valuable resource for understanding consumer health-related posts on social media. We benchmark the dataset on multiple state-of-the-art summarization models to show the effectiveness of the dataset
Lessons from the TREC Plain Language Adaptation of Biomedical Abstracts (PLABA) track
Jul 18, 2025
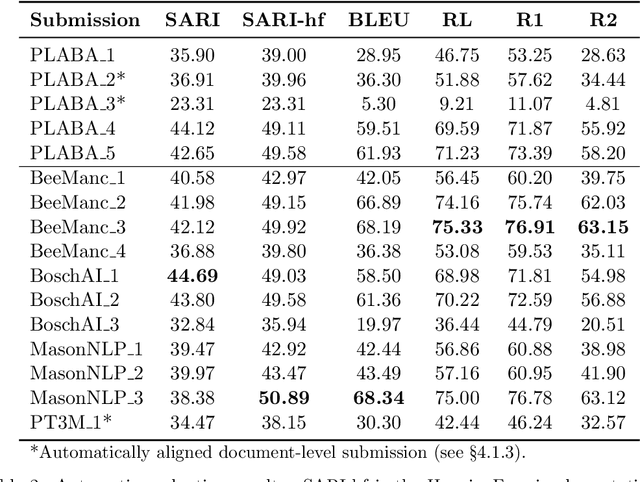
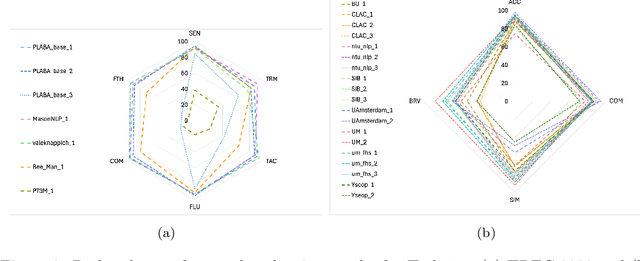
Abstract:Objective: Recent advances in language models have shown potential to adapt professional-facing biomedical literature to plain language, making it accessible to patients and caregivers. However, their unpredictability, combined with the high potential for harm in this domain, means rigorous evaluation is necessary. Our goals with this track were to stimulate research and to provide high-quality evaluation of the most promising systems. Methods: We hosted the Plain Language Adaptation of Biomedical Abstracts (PLABA) track at the 2023 and 2024 Text Retrieval Conferences. Tasks included complete, sentence-level, rewriting of abstracts (Task 1) as well as identifying and replacing difficult terms (Task 2). For automatic evaluation of Task 1, we developed a four-fold set of professionally-written references. Submissions for both Tasks 1 and 2 were provided extensive manual evaluation from biomedical experts. Results: Twelve teams spanning twelve countries participated in the track, with models from multilayer perceptrons to large pretrained transformers. In manual judgments of Task 1, top-performing models rivaled human levels of factual accuracy and completeness, but not simplicity or brevity. Automatic, reference-based metrics generally did not correlate well with manual judgments. In Task 2, systems struggled with identifying difficult terms and classifying how to replace them. When generating replacements, however, LLM-based systems did well in manually judged accuracy, completeness, and simplicity, though not in brevity. Conclusion: The PLABA track showed promise for using Large Language Models to adapt biomedical literature for the general public, while also highlighting their deficiencies and the need for improved automatic benchmarking tools.
JEBS: A Fine-grained Biomedical Lexical Simplification Task
Jun 15, 2025



Abstract:Online medical literature has made health information more available than ever, however, the barrier of complex medical jargon prevents the general public from understanding it. Though parallel and comparable corpora for Biomedical Text Simplification have been introduced, these conflate the many syntactic and lexical operations involved in simplification. To enable more targeted development and evaluation, we present a fine-grained lexical simplification task and dataset, Jargon Explanations for Biomedical Simplification (JEBS, https://github.com/bill-from-ri/JEBS-data ). The JEBS task involves identifying complex terms, classifying how to replace them, and generating replacement text. The JEBS dataset contains 21,595 replacements for 10,314 terms across 400 biomedical abstracts and their manually simplified versions. Additionally, we provide baseline results for a variety of rule-based and transformer-based systems for the three sub-tasks. The JEBS task, data, and baseline results pave the way for development and rigorous evaluation of systems for replacing or explaining complex biomedical terms.
A Dataset for Addressing Patient's Information Needs related to Clinical Course of Hospitalization
Jun 04, 2025Abstract:Patients have distinct information needs about their hospitalization that can be addressed using clinical evidence from electronic health records (EHRs). While artificial intelligence (AI) systems show promise in meeting these needs, robust datasets are needed to evaluate the factual accuracy and relevance of AI-generated responses. To our knowledge, no existing dataset captures patient information needs in the context of their EHRs. We introduce ArchEHR-QA, an expert-annotated dataset based on real-world patient cases from intensive care unit and emergency department settings. The cases comprise questions posed by patients to public health forums, clinician-interpreted counterparts, relevant clinical note excerpts with sentence-level relevance annotations, and clinician-authored answers. To establish benchmarks for grounded EHR question answering (QA), we evaluated three open-weight large language models (LLMs)--Llama 4, Llama 3, and Mixtral--across three prompting strategies: generating (1) answers with citations to clinical note sentences, (2) answers before citations, and (3) answers from filtered citations. We assessed performance on two dimensions: Factuality (overlap between cited note sentences and ground truth) and Relevance (textual and semantic similarity between system and reference answers). The final dataset contains 134 patient cases. The answer-first prompting approach consistently performed best, with Llama 4 achieving the highest scores. Manual error analysis supported these findings and revealed common issues such as omitted key clinical evidence and contradictory or hallucinated content. Overall, ArchEHR-QA provides a strong benchmark for developing and evaluating patient-centered EHR QA systems, underscoring the need for further progress toward generating factual and relevant responses in clinical contexts.
Overview of TREC 2024 Medical Video Question Answering (MedVidQA) Track
Dec 15, 2024



Abstract:One of the key goals of artificial intelligence (AI) is the development of a multimodal system that facilitates communication with the visual world (image and video) using a natural language query. Earlier works on medical question answering primarily focused on textual and visual (image) modalities, which may be inefficient in answering questions requiring demonstration. In recent years, significant progress has been achieved due to the introduction of large-scale language-vision datasets and the development of efficient deep neural techniques that bridge the gap between language and visual understanding. Improvements have been made in numerous vision-and-language tasks, such as visual captioning visual question answering, and natural language video localization. Most of the existing work on language vision focused on creating datasets and developing solutions for open-domain applications. We believe medical videos may provide the best possible answers to many first aid, medical emergency, and medical education questions. With increasing interest in AI to support clinical decision-making and improve patient engagement, there is a need to explore such challenges and develop efficient algorithms for medical language-video understanding and generation. Toward this, we introduced new tasks to foster research toward designing systems that can understand medical videos to provide visual answers to natural language questions, and are equipped with multimodal capability to generate instruction steps from the medical video. These tasks have the potential to support the development of sophisticated downstream applications that can benefit the public and medical professionals.
Overview of TREC 2024 Biomedical Generative Retrieval (BioGen) Track
Nov 27, 2024



Abstract:With the advancement of large language models (LLMs), the biomedical domain has seen significant progress and improvement in multiple tasks such as biomedical question answering, lay language summarization of the biomedical literature, clinical note summarization, etc. However, hallucinations or confabulations remain one of the key challenges when using LLMs in the biomedical and other domains. Inaccuracies may be particularly harmful in high-risk situations, such as making clinical decisions or appraising biomedical research. Studies on the evaluation of the LLMs' abilities to ground generated statements in verifiable sources have shown that models perform significantly worse on lay-user generated questions, and often fail to reference relevant sources. This can be problematic when those seeking information want evidence from studies to back up the claims from LLMs[3]. Unsupported statements are a major barrier to using LLMs in any applications that may affect health. Methods for grounding generated statements in reliable sources along with practical evaluation approaches are needed to overcome this barrier. Towards this, in our pilot task organized at TREC 2024, we introduced the task of reference attribution as a means to mitigate the generation of false statements by LLMs answering biomedical questions.
Toward Relieving Clinician Burden by Automatically Generating Progress Notes using Interim Hospital Data
Oct 10, 2024



Abstract:Regular documentation of progress notes is one of the main contributors to clinician burden. The abundance of structured chart information in medical records further exacerbates the burden, however, it also presents an opportunity to automate the generation of progress notes. In this paper, we propose a task to automate progress note generation using structured or tabular information present in electronic health records. To this end, we present a novel framework and a large dataset, ChartPNG, for the task which contains $7089$ annotation instances (each having a pair of progress notes and interim structured chart data) across $1616$ patients. We establish baselines on the dataset using large language models from general and biomedical domains. We perform both automated (where the best performing Biomistral model achieved a BERTScore F1 of $80.53$ and MEDCON score of $19.61$) and manual (where we found that the model was able to leverage relevant structured data with $76.9\%$ accuracy) analyses to identify the challenges with the proposed task and opportunities for future research.
Towards Answering Health-related Questions from Medical Videos: Datasets and Approaches
Sep 21, 2023



Abstract:The increase in the availability of online videos has transformed the way we access information and knowledge. A growing number of individuals now prefer instructional videos as they offer a series of step-by-step procedures to accomplish particular tasks. The instructional videos from the medical domain may provide the best possible visual answers to first aid, medical emergency, and medical education questions. Toward this, this paper is focused on answering health-related questions asked by the public by providing visual answers from medical videos. The scarcity of large-scale datasets in the medical domain is a key challenge that hinders the development of applications that can help the public with their health-related questions. To address this issue, we first proposed a pipelined approach to create two large-scale datasets: HealthVidQA-CRF and HealthVidQA-Prompt. Later, we proposed monomodal and multimodal approaches that can effectively provide visual answers from medical videos to natural language questions. We conducted a comprehensive analysis of the results, focusing on the impact of the created datasets on model training and the significance of visual features in enhancing the performance of the monomodal and multi-modal approaches. Our findings suggest that these datasets have the potential to enhance the performance of medical visual answer localization tasks and provide a promising future direction to further enhance the performance by using pre-trained language-vision models.
Empowering Language Model with Guided Knowledge Fusion for Biomedical Document Re-ranking
May 07, 2023



Abstract:Pre-trained language models (PLMs) have proven to be effective for document re-ranking task. However, they lack the ability to fully interpret the semantics of biomedical and health-care queries and often rely on simplistic patterns for retrieving documents. To address this challenge, we propose an approach that integrates knowledge and the PLMs to guide the model toward effectively capturing information from external sources and retrieving the correct documents. We performed comprehensive experiments on two biomedical and open-domain datasets that show that our approach significantly improves vanilla PLMs and other existing approaches for document re-ranking task.
Medical Image Retrieval via Nearest Neighbor Search on Pre-trained Image Features
Oct 05, 2022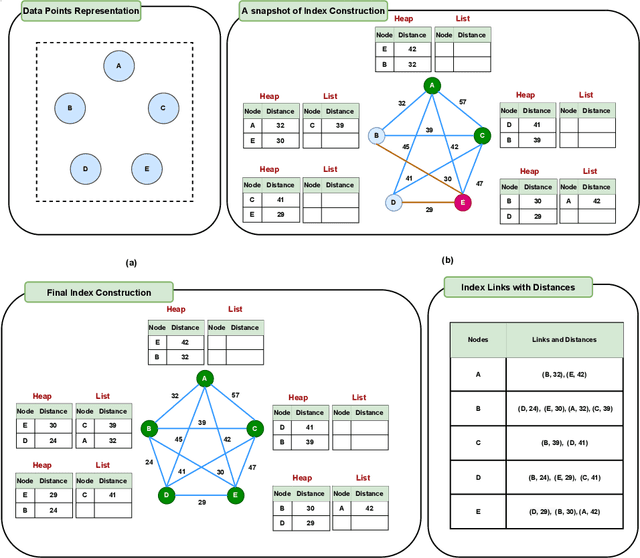

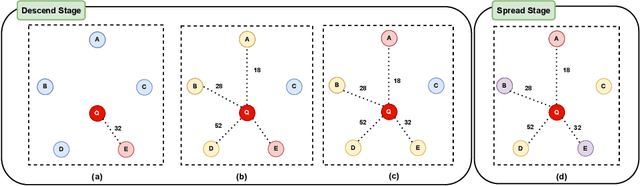
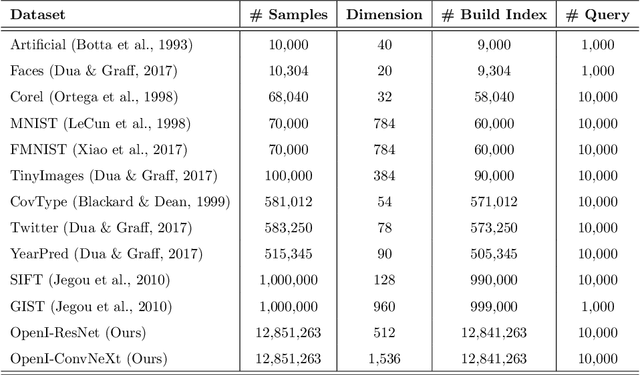
Abstract:Nearest neighbor search (NNS) aims to locate the points in high-dimensional space that is closest to the query point. The brute-force approach for finding the nearest neighbor becomes computationally infeasible when the number of points is large. The NNS has multiple applications in medicine, such as searching large medical imaging databases, disease classification, diagnosis, etc. With a focus on medical imaging, this paper proposes DenseLinkSearch an effective and efficient algorithm that searches and retrieves the relevant images from heterogeneous sources of medical images. Towards this, given a medical database, the proposed algorithm builds the index that consists of pre-computed links of each point in the database. The search algorithm utilizes the index to efficiently traverse the database in search of the nearest neighbor. We extensively tested the proposed NNS approach and compared the performance with state-of-the-art NNS approaches on benchmark datasets and our created medical image datasets. The proposed approach outperformed the existing approach in terms of retrieving accurate neighbors and retrieval speed. We also explore the role of medical image feature representation in content-based medical image retrieval tasks. We propose a Transformer-based feature representation technique that outperformed the existing pre-trained Transformer approach on CLEF 2011 medical image retrieval task. The source code of our experiments are available at https://github.com/deepaknlp/DLS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge