Sarvesh Soni
A Dataset for Addressing Patient's Information Needs related to Clinical Course of Hospitalization
Jun 04, 2025Abstract:Patients have distinct information needs about their hospitalization that can be addressed using clinical evidence from electronic health records (EHRs). While artificial intelligence (AI) systems show promise in meeting these needs, robust datasets are needed to evaluate the factual accuracy and relevance of AI-generated responses. To our knowledge, no existing dataset captures patient information needs in the context of their EHRs. We introduce ArchEHR-QA, an expert-annotated dataset based on real-world patient cases from intensive care unit and emergency department settings. The cases comprise questions posed by patients to public health forums, clinician-interpreted counterparts, relevant clinical note excerpts with sentence-level relevance annotations, and clinician-authored answers. To establish benchmarks for grounded EHR question answering (QA), we evaluated three open-weight large language models (LLMs)--Llama 4, Llama 3, and Mixtral--across three prompting strategies: generating (1) answers with citations to clinical note sentences, (2) answers before citations, and (3) answers from filtered citations. We assessed performance on two dimensions: Factuality (overlap between cited note sentences and ground truth) and Relevance (textual and semantic similarity between system and reference answers). The final dataset contains 134 patient cases. The answer-first prompting approach consistently performed best, with Llama 4 achieving the highest scores. Manual error analysis supported these findings and revealed common issues such as omitted key clinical evidence and contradictory or hallucinated content. Overall, ArchEHR-QA provides a strong benchmark for developing and evaluating patient-centered EHR QA systems, underscoring the need for further progress toward generating factual and relevant responses in clinical contexts.
Toward Relieving Clinician Burden by Automatically Generating Progress Notes using Interim Hospital Data
Oct 10, 2024



Abstract:Regular documentation of progress notes is one of the main contributors to clinician burden. The abundance of structured chart information in medical records further exacerbates the burden, however, it also presents an opportunity to automate the generation of progress notes. In this paper, we propose a task to automate progress note generation using structured or tabular information present in electronic health records. To this end, we present a novel framework and a large dataset, ChartPNG, for the task which contains $7089$ annotation instances (each having a pair of progress notes and interim structured chart data) across $1616$ patients. We establish baselines on the dataset using large language models from general and biomedical domains. We perform both automated (where the best performing Biomistral model achieved a BERTScore F1 of $80.53$ and MEDCON score of $19.61$) and manual (where we found that the model was able to leverage relevant structured data with $76.9\%$ accuracy) analyses to identify the challenges with the proposed task and opportunities for future research.
Toward a Neural Semantic Parsing System for EHR Question Answering
Nov 08, 2022Abstract:Clinical semantic parsing (SP) is an important step toward identifying the exact information need (as a machine-understandable logical form) from a natural language query aimed at retrieving information from electronic health records (EHRs). Current approaches to clinical SP are largely based on traditional machine learning and require hand-building a lexicon. The recent advancements in neural SP show a promise for building a robust and flexible semantic parser without much human effort. Thus, in this paper, we aim to systematically assess the performance of two such neural SP models for EHR question answering (QA). We found that the performance of these advanced neural models on two clinical SP datasets is promising given their ease of application and generalizability. Our error analysis surfaces the common types of errors made by these models and has the potential to inform future research into improving the performance of neural SP models for EHR QA.
Patient Cohort Retrieval using Transformer Language Models
Sep 10, 2020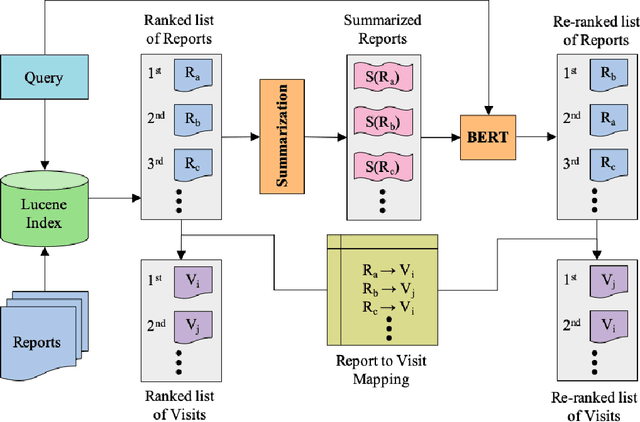

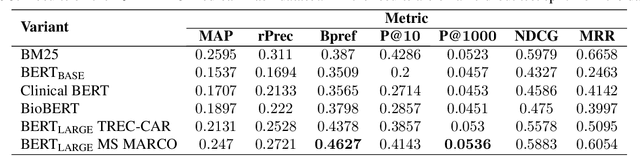
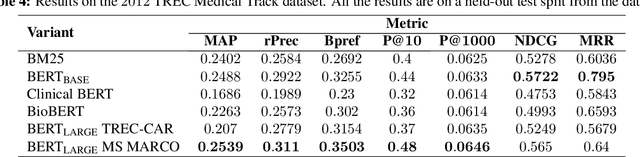
Abstract:We apply deep learning-based language models to the task of patient cohort retrieval (CR) with the aim to assess their efficacy. The task of CR requires the extraction of relevant documents from the electronic health records (EHRs) on the basis of a given query. Given the recent advancements in the field of document retrieval, we map the task of CR to a document retrieval task and apply various deep neural models implemented for the general domain tasks. In this paper, we propose a framework for retrieving patient cohorts using neural language models without the need of explicit feature engineering and domain expertise. We find that a majority of our models outperform the BM25 baseline method on various evaluation metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge