Cristian Regep
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024



Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
RECOVER: sequential model optimization platform for combination drug repurposing identifies novel synergistic compounds in vitro
Feb 07, 2022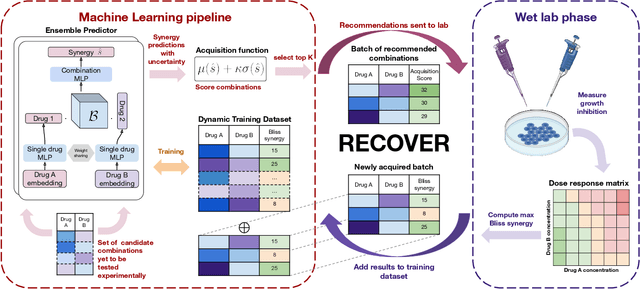
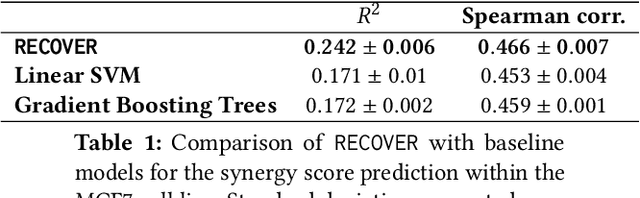
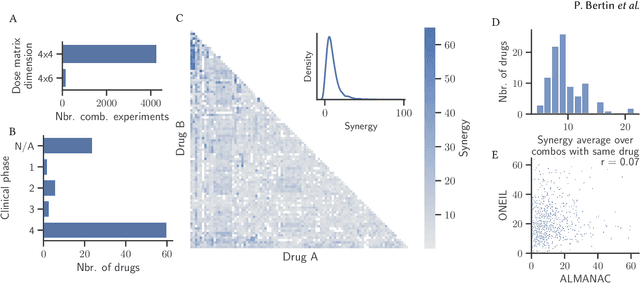
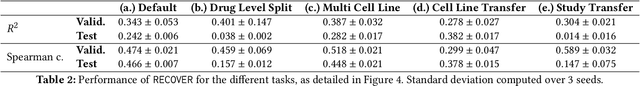
Abstract:Selecting optimal drug repurposing combinations for further preclinical development is a challenging technical feat. Due to the toxicity of many therapeutic agents (e.g., chemotherapy), practitioners have favoured selection of synergistic compounds whereby lower doses can be used whilst maintaining high efficacy. For a fixed small molecule library, an exhaustive combinatorial chemical screen becomes infeasible to perform for academic and industry laboratories alike. Deep learning models have achieved state-of-the-art results in silico for the prediction of synergy scores. However, databases of drug combinations are highly biased towards synergistic agents and these results do not necessarily generalise out of distribution. We employ a sequential model optimization search applied to a deep learning model to quickly discover highly synergistic drug combinations active against a cancer cell line, while requiring substantially less screening than an exhaustive evaluation. Through iteratively adapting the model to newly acquired data, after only 3 rounds of ML-guided experimentation (including a calibration round), we find that the set of combinations queried by our model is enriched for highly synergistic combinations. Remarkably, we rediscovered a synergistic drug combination that was later confirmed to be under study within clinical trials.
Utilising Graph Machine Learning within Drug Discovery and Development
Dec 09, 2020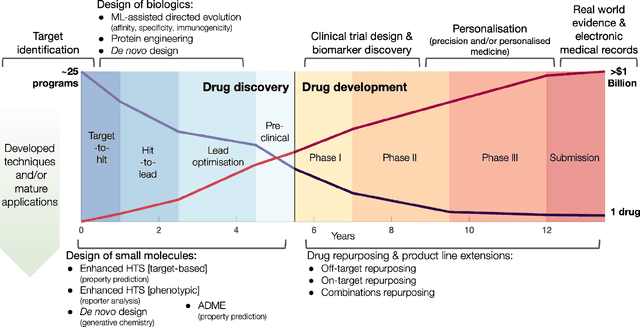
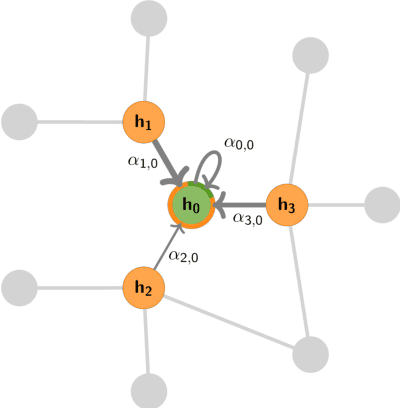
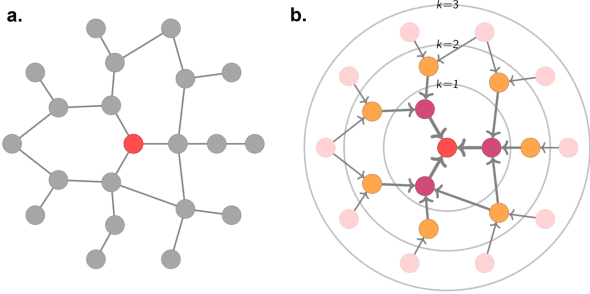
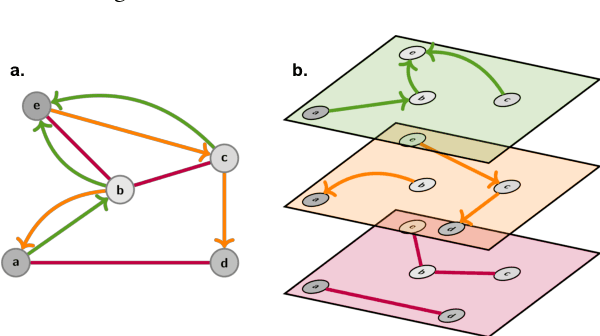
Abstract:Graph Machine Learning (GML) is receiving growing interest within the pharmaceutical and biotechnology industries for its ability to model biomolecular structures, the functional relationships between them, and integrate multi-omic datasets - amongst other data types. Herein, we present a multidisciplinary academic-industrial review of the topic within the context of drug discovery and development. After introducing key terms and modelling approaches, we move chronologically through the drug development pipeline to identify and summarise work incorporating: target identification, design of small molecules and biologics, and drug repurposing. Whilst the field is still emerging, key milestones including repurposed drugs entering in vivo studies, suggest graph machine learning will become a modelling framework of choice within biomedical machine learning.
Sparse Dynamic Distribution Decomposition: Efficient Integration of Trajectory and Snapshot Time Series Data
Jun 11, 2020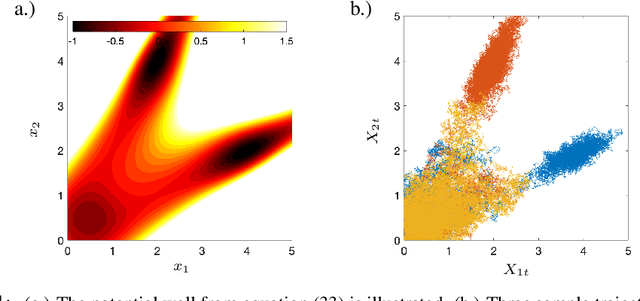
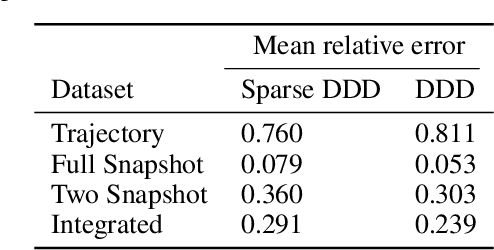
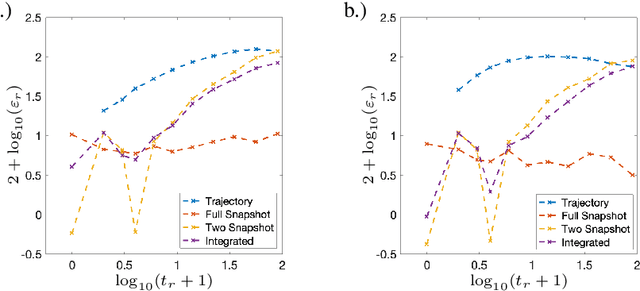
Abstract:Dynamic Distribution Decomposition (DDD) was introduced in Taylor-King et. al. (PLOS Comp Biol, 2020) as a variation on Dynamic Mode Decomposition. In brief, by using basis functions over a continuous state space, DDD allows for the fitting of continuous-time Markov chains over these basis functions and as a result continuously maps between distributions. The number of parameters in DDD scales by the square of the number of basis functions; we reformulate the problem and restrict the method to compact basis functions which leads to the inference of sparse matrices only -- hence reducing the number of parameters. Finally, we demonstrate how DDD is suitable to integrate both trajectory time series (paired between subsequent time points) and snapshot time series (unpaired time points). Methods capable of integrating both scenarios are particularly relevant for the analysis of biomedical data, whereby studies observe population at fixed time points (snapshots) and individual patient journeys with repeated follow ups (trajectories).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge