Byron C. Wallace
Biomedical Interpretable Entity Representations
Jun 17, 2021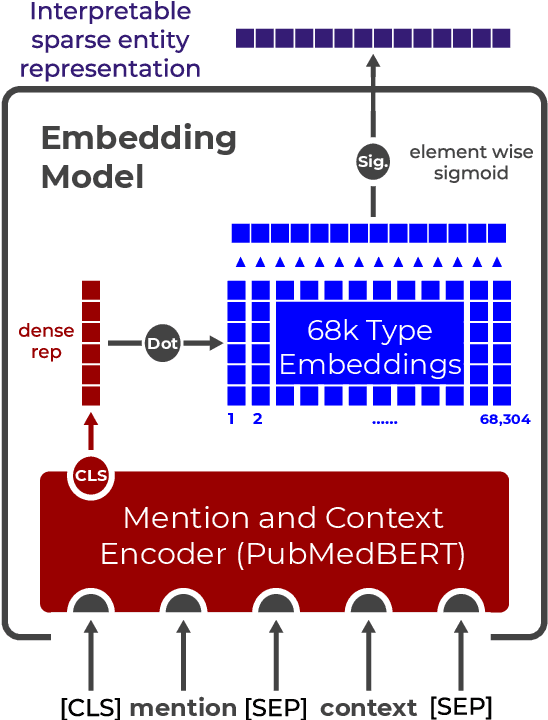
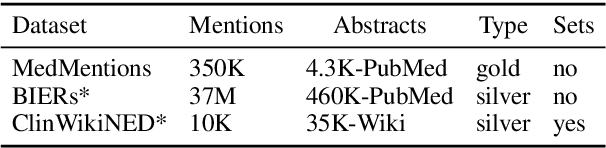
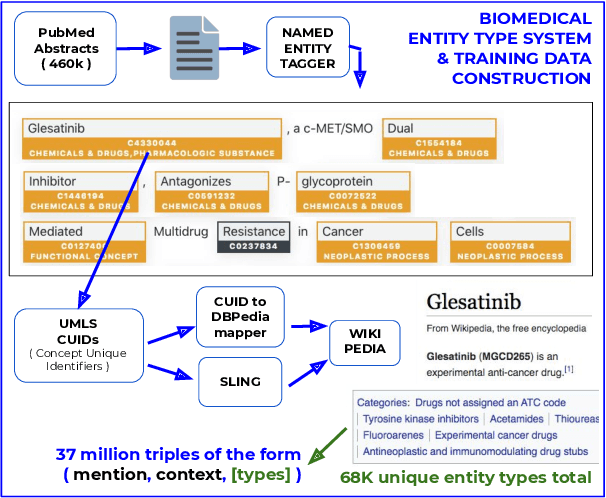
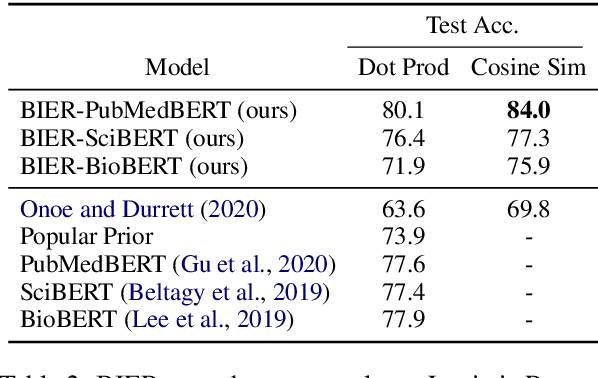
Abstract:Pre-trained language models induce dense entity representations that offer strong performance on entity-centric NLP tasks, but such representations are not immediately interpretable. This can be a barrier to model uptake in important domains such as biomedicine. There has been recent work on general interpretable representation learning (Onoe and Durrett, 2020), but these domain-agnostic representations do not readily transfer to the important domain of biomedicine. In this paper, we create a new entity type system and training set from a large corpus of biomedical texts by mapping entities to concepts in a medical ontology, and from these to Wikipedia pages whose categories are our types. From this mapping we derive Biomedical Interpretable Entity Representations(BIERs), in which dimensions correspond to fine-grained entity types, and values are predicted probabilities that a given entity is of the corresponding type. We propose a novel method that exploits BIER's final sparse and intermediate dense representations to facilitate model and entity type debugging. We show that BIERs achieve strong performance in biomedical tasks including named entity disambiguation and entity label classification, and we provide error analysis to highlight the utility of their interpretability, particularly in low-supervision settings. Finally, we provide our induced 68K biomedical type system, the corresponding 37 million triples of derived data used to train BIER models and our best performing model.
Does BERT Pretrained on Clinical Notes Reveal Sensitive Data?
Apr 22, 2021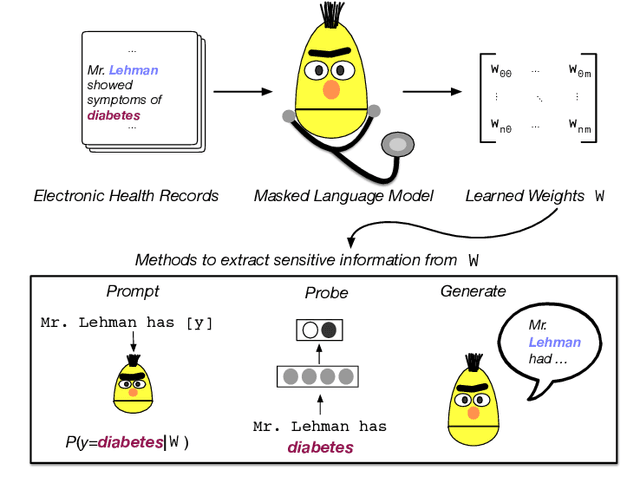

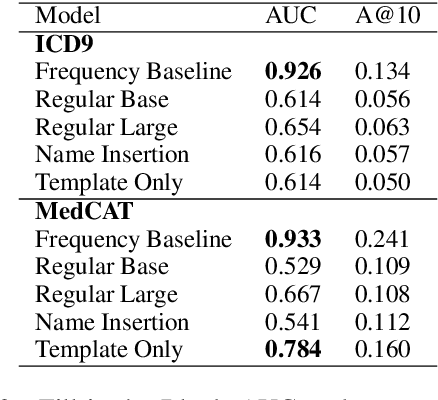
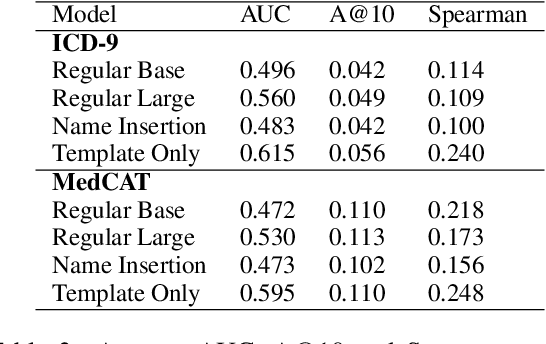
Abstract:Large Transformers pretrained over clinical notes from Electronic Health Records (EHR) have afforded substantial gains in performance on predictive clinical tasks. The cost of training such models (and the necessity of data access to do so) coupled with their utility motivates parameter sharing, i.e., the release of pretrained models such as ClinicalBERT. While most efforts have used deidentified EHR, many researchers have access to large sets of sensitive, non-deidentified EHR with which they might train a BERT model (or similar). Would it be safe to release the weights of such a model if they did? In this work, we design a battery of approaches intended to recover Personal Health Information (PHI) from a trained BERT. Specifically, we attempt to recover patient names and conditions with which they are associated. We find that simple probing methods are not able to meaningfully extract sensitive information from BERT trained over the MIMIC-III corpus of EHR. However, more sophisticated "attacks" may succeed in doing so: To facilitate such research, we make our experimental setup and baseline probing models available at https://github.com/elehman16/exposing_patient_data_release
Disentangling Representations of Text by Masking Transformers
Apr 14, 2021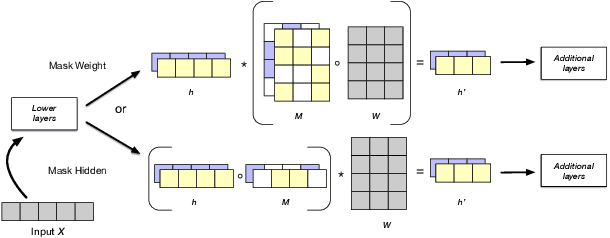
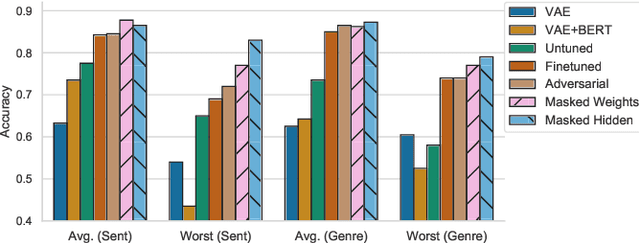
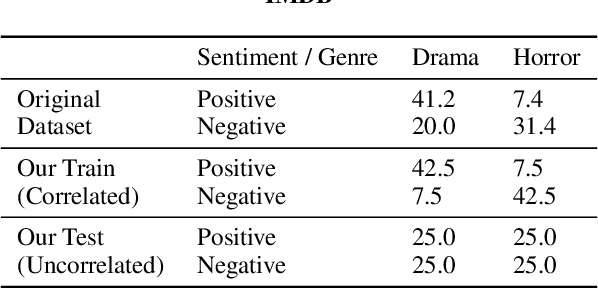
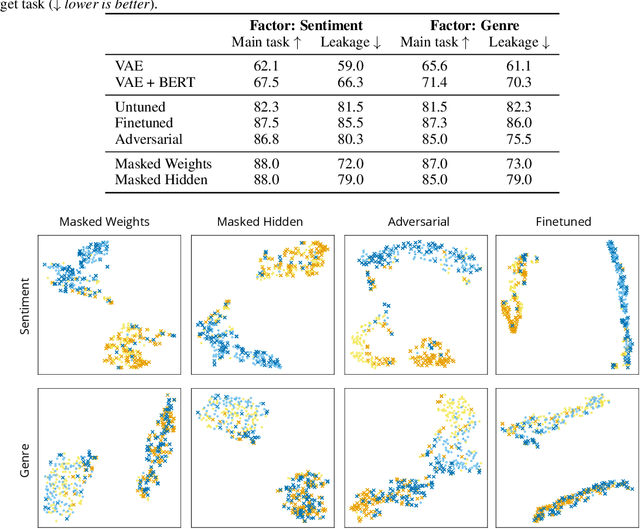
Abstract:Representations from large pretrained models such as BERT encode a range of features into monolithic vectors, affording strong predictive accuracy across a multitude of downstream tasks. In this paper we explore whether it is possible to learn disentangled representations by identifying existing subnetworks within pretrained models that encode distinct, complementary aspect representations. Concretely, we learn binary masks over transformer weights or hidden units to uncover subsets of features that correlate with a specific factor of variation; this eliminates the need to train a disentangled model from scratch for a particular task. We evaluate this method with respect to its ability to disentangle representations of sentiment from genre in movie reviews, "toxicity" from dialect in Tweets, and syntax from semantics. By combining masking with magnitude pruning we find that we can identify sparse subnetworks within BERT that strongly encode particular aspects (e.g., toxicity) while only weakly encoding others (e.g., race). Moreover, despite only learning masks, we find that disentanglement-via-masking performs as well as -- and often better than -- previously proposed methods based on variational autoencoders and adversarial training.
On the Impact of Random Seeds on the Fairness of Clinical Classifiers
Apr 13, 2021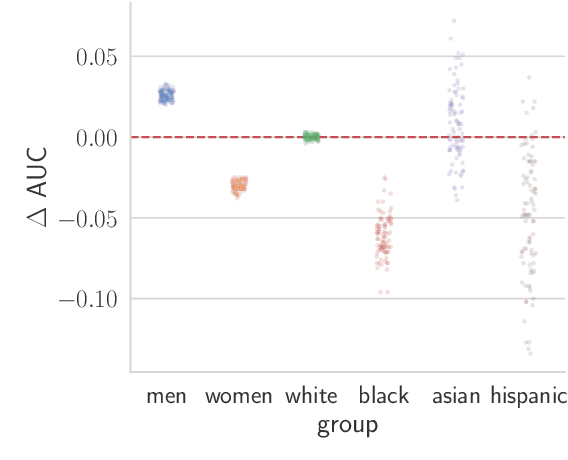
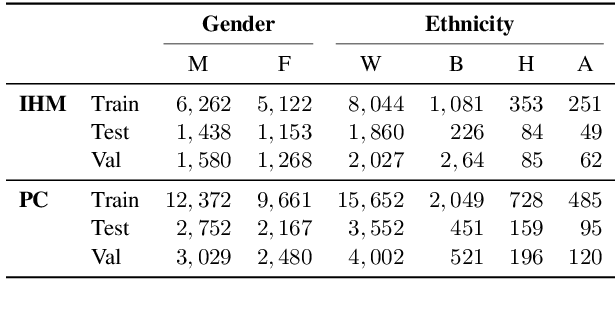
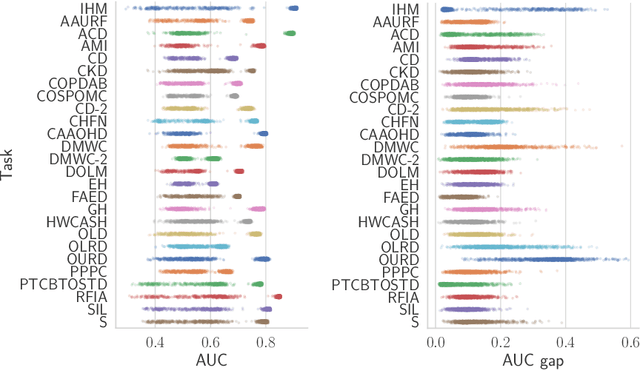
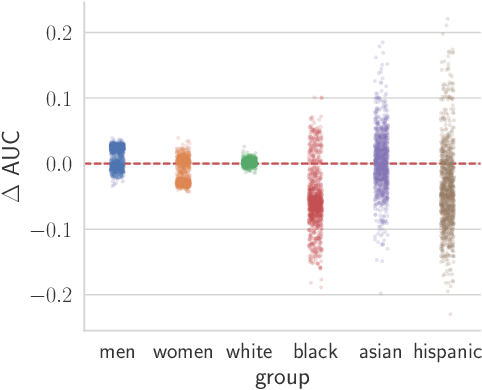
Abstract:Recent work has shown that fine-tuning large networks is surprisingly sensitive to changes in random seed(s). We explore the implications of this phenomenon for model fairness across demographic groups in clinical prediction tasks over electronic health records (EHR) in MIMIC-III -- the standard dataset in clinical NLP research. Apparent subgroup performance varies substantially for seeds that yield similar overall performance, although there is no evidence of a trade-off between overall and subgroup performance. However, we also find that the small sample sizes inherent to looking at intersections of minority groups and somewhat rare conditions limit our ability to accurately estimate disparities. Further, we find that jointly optimizing for high overall performance and low disparities does not yield statistically significant improvements. Our results suggest that fairness work using MIMIC-III should carefully account for variations in apparent differences that may arise from stochasticity and small sample sizes.
Paragraph-level Simplification of Medical Texts
Apr 12, 2021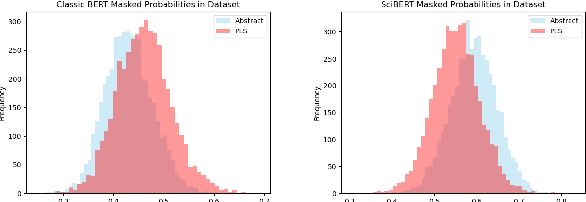
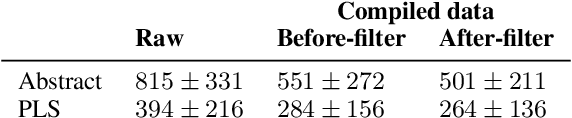
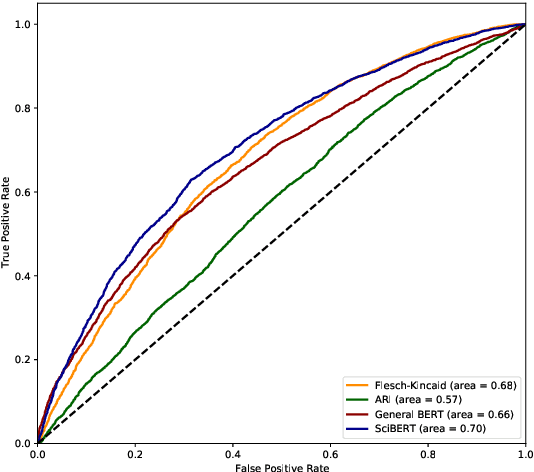

Abstract:We consider the problem of learning to simplify medical texts. This is important because most reliable, up-to-date information in biomedicine is dense with jargon and thus practically inaccessible to the lay audience. Furthermore, manual simplification does not scale to the rapidly growing body of biomedical literature, motivating the need for automated approaches. Unfortunately, there are no large-scale resources available for this task. In this work we introduce a new corpus of parallel texts in English comprising technical and lay summaries of all published evidence pertaining to different clinical topics. We then propose a new metric based on likelihood scores from a masked language model pretrained on scientific texts. We show that this automated measure better differentiates between technical and lay summaries than existing heuristics. We introduce and evaluate baseline encoder-decoder Transformer models for simplification and propose a novel augmentation to these in which we explicitly penalize the decoder for producing "jargon" terms; we find that this yields improvements over baselines in terms of readability.
An Empirical Comparison of Instance Attribution Methods for NLP
Apr 09, 2021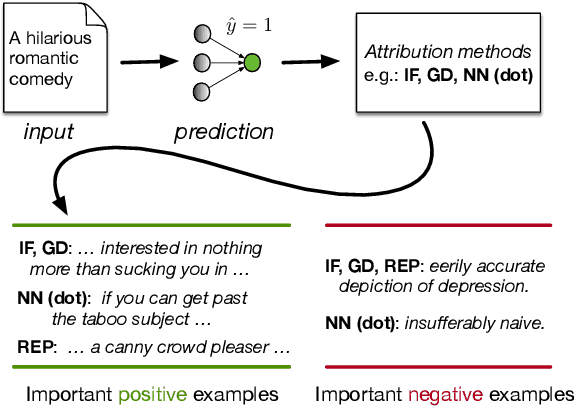
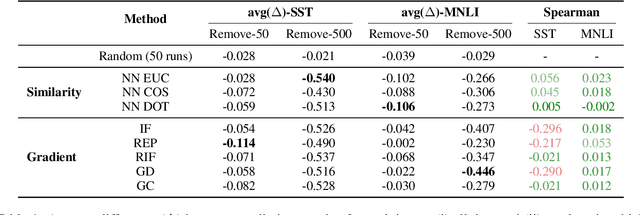
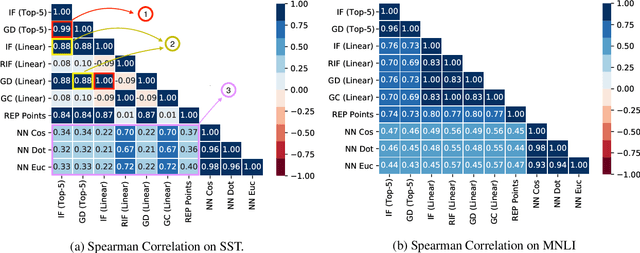
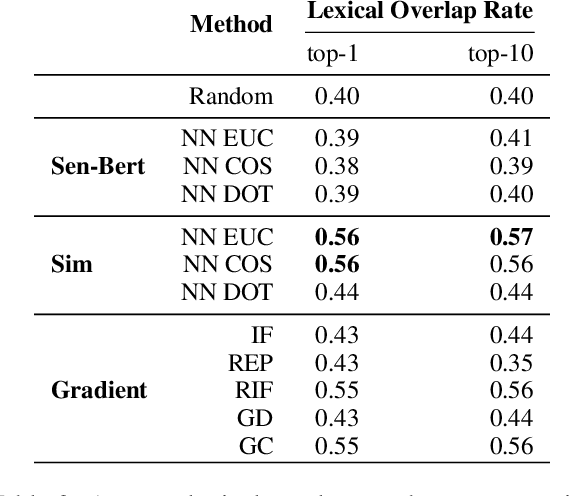
Abstract:Widespread adoption of deep models has motivated a pressing need for approaches to interpret network outputs and to facilitate model debugging. Instance attribution methods constitute one means of accomplishing these goals by retrieving training instances that (may have) led to a particular prediction. Influence functions (IF; Koh and Liang 2017) provide machinery for doing this by quantifying the effect that perturbing individual train instances would have on a specific test prediction. However, even approximating the IF is computationally expensive, to the degree that may be prohibitive in many cases. Might simpler approaches (e.g., retrieving train examples most similar to a given test point) perform comparably? In this work, we evaluate the degree to which different potential instance attribution agree with respect to the importance of training samples. We find that simple retrieval methods yield training instances that differ from those identified via gradient-based methods (such as IFs), but that nonetheless exhibit desirable characteristics similar to more complex attribution methods. Code for all methods and experiments in this paper is available at: https://github.com/successar/instance_attributions_NLP.
Unsupervised Data Augmentation with Naive Augmentation and without Unlabeled Data
Oct 22, 2020
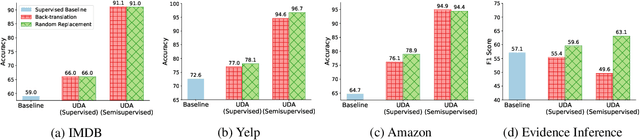
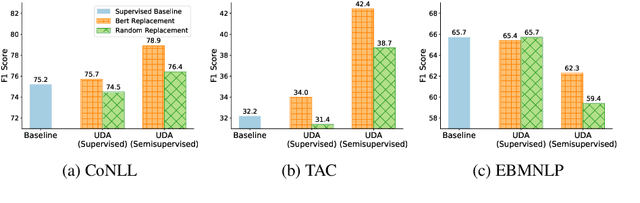

Abstract:Unsupervised Data Augmentation (UDA) is a semi-supervised technique that applies a consistency loss to penalize differences between a model's predictions on (a) observed (unlabeled) examples; and (b) corresponding 'noised' examples produced via data augmentation. While UDA has gained popularity for text classification, open questions linger over which design decisions are necessary and over how to extend the method to sequence labeling tasks. This method has recently gained traction for text classification. In this paper, we re-examine UDA and demonstrate its efficacy on several sequential tasks. Our main contribution is an empirical study of UDA to establish which components of the algorithm confer benefits in NLP. Notably, although prior work has emphasized the use of clever augmentation techniques including back-translation, we find that enforcing consistency between predictions assigned to observed and randomly substituted words often yields comparable (or greater) benefits compared to these complex perturbation models. Furthermore, we find that applying its consistency loss affords meaningful gains without any unlabeled data at all, i.e., in a standard supervised setting. In short: UDA need not be unsupervised, and does not require complex data augmentation to be effective.
Understanding Clinical Trial Reports: Extracting Medical Entities and Their Relations
Oct 08, 2020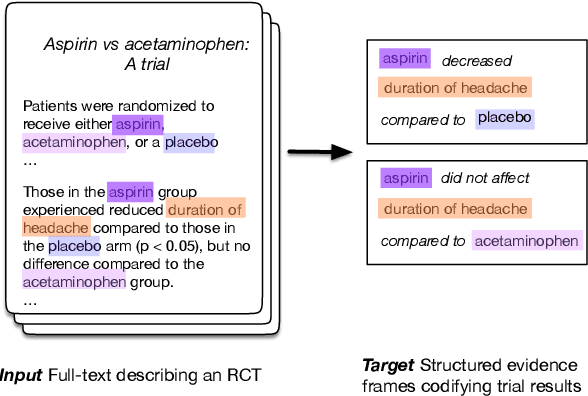
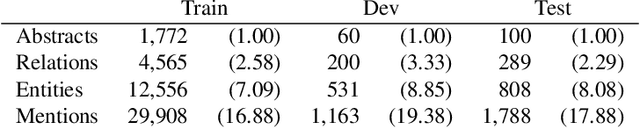
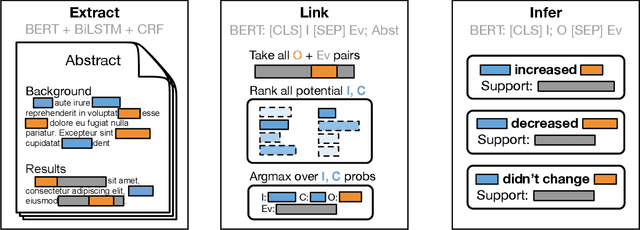

Abstract:The best evidence concerning comparative treatment effectiveness comes from clinical trials, the results of which are reported in unstructured articles. Medical experts must manually extract information from articles to inform decision-making, which is time-consuming and expensive. Here we consider the end-to-end task of both (a) extracting treatments and outcomes from full-text articles describing clinical trials (entity identification) and, (b) inferring the reported results for the former with respect to the latter (relation extraction). We introduce new data for this task, and evaluate models that have recently achieved state-of-the-art results on similar tasks in Natural Language Processing. We then propose a new method motivated by how trial results are typically presented that outperforms these purely data-driven baselines. Finally, we run a fielded evaluation of the model with a non-profit seeking to identify existing drugs that might be re-purposed for cancer, showing the potential utility of end-to-end evidence extraction systems.
Generating (Factual?) Narrative Summaries of RCTs: Experiments with Neural Multi-Document Summarization
Aug 25, 2020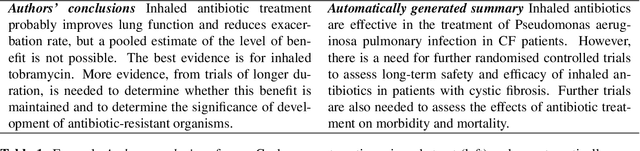
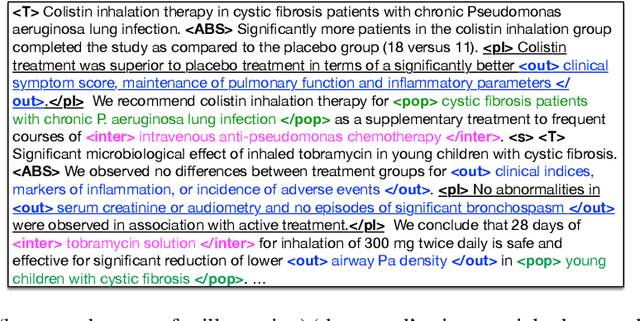

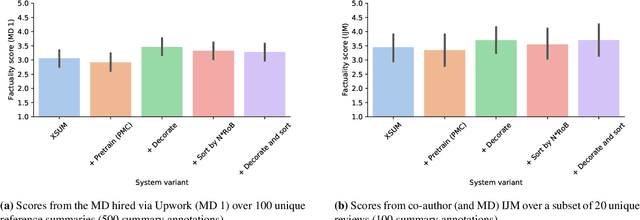
Abstract:We consider the problem of automatically generating a narrative biomedical evidence summary from multiple trial reports. We evaluate modern neural models for abstractive summarization of relevant article abstracts from systematic reviews previously conducted by members of the Cochrane collaboration, using the authors conclusions section of the review abstract as our target. We enlist medical professionals to evaluate generated summaries, and we find that modern summarization systems yield consistently fluent and relevant synopses, but that they are not always factual. We propose new approaches that capitalize on domain-specific models to inform summarization, e.g., by explicitly demarcating snippets of inputs that convey key findings, and emphasizing the reports of large and high-quality trials. We find that these strategies modestly improve the factual accuracy of generated summaries. Finally, we propose a new method for automatically evaluating the factuality of generated narrative evidence syntheses using models that infer the directionality of reported findings.
Semi-Automating Knowledge Base Construction for Cancer Genetics
May 26, 2020
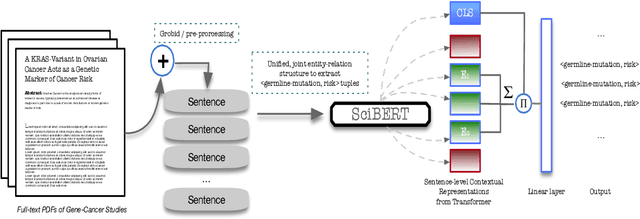
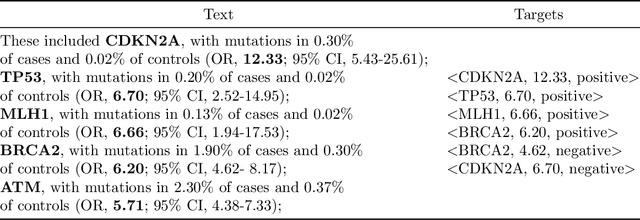

Abstract:In this work, we consider the exponentially growing subarea of genetics in cancer. The need to synthesize and centralize this evidence for dissemination has motivated a team of physicians to manually construct and maintain a knowledge base that distills key results reported in the literature. This is a laborious process that entails reading through full-text articles to understand the study design, assess study quality, and extract the reported cancer risk estimates associated with particular hereditary cancer genes (i.e., penetrance). In this work, we propose models to automatically surface key elements from full-text cancer genetics articles, with the ultimate aim of expediting the manual workflow currently in place. We propose two challenging tasks that are critical for characterizing the findings reported cancer genetics studies: (i) Extracting snippets of text that describe \emph{ascertainment mechanisms}, which in turn inform whether the population studied may introduce bias owing to deviations from the target population; (ii) Extracting reported risk estimates (e.g., odds or hazard ratios) associated with specific germline mutations. The latter task may be viewed as a joint entity tagging and relation extraction problem. To train models for these tasks, we induce distant supervision over tokens and snippets in full-text articles using the manually constructed knowledge base. We propose and evaluate several model variants, including a transformer-based joint entity and relation extraction model to extract <germline mutation, risk-estimate>} pairs. We observe strong empirical performance, highlighting the practical potential for such models to aid KB construction in this space. We ablate components of our model, observing, e.g., that a joint model for <germline mutation, risk-estimate> fares substantially better than a pipelined approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge