Neuro-Causal Factor Analysis
May 31, 2023Alex Markham, Mingyu Liu, Bryon Aragam, Liam Solus
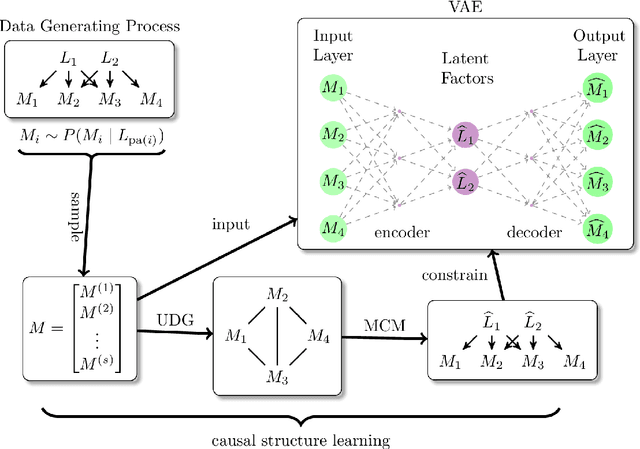
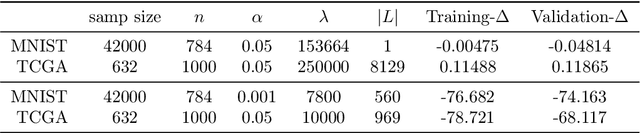
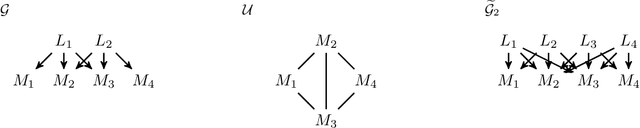

Factor analysis (FA) is a statistical tool for studying how observed variables with some mutual dependences can be expressed as functions of mutually independent unobserved factors, and it is widely applied throughout the psychological, biological, and physical sciences. We revisit this classic method from the comparatively new perspective given by advancements in causal discovery and deep learning, introducing a framework for Neuro-Causal Factor Analysis (NCFA). Our approach is fully nonparametric: it identifies factors via latent causal discovery methods and then uses a variational autoencoder (VAE) that is constrained to abide by the Markov factorization of the distribution with respect to the learned graph. We evaluate NCFA on real and synthetic data sets, finding that it performs comparably to standard VAEs on data reconstruction tasks but with the advantages of sparser architecture, lower model complexity, and causal interpretability. Unlike traditional FA methods, our proposed NCFA method allows learning and reasoning about the latent factors underlying observed data from a justifiably causal perspective, even when the relations between factors and measurements are highly nonlinear.
Optimizing NOTEARS Objectives via Topological Swaps
May 26, 2023Chang Deng, Kevin Bello, Bryon Aragam, Pradeep Ravikumar
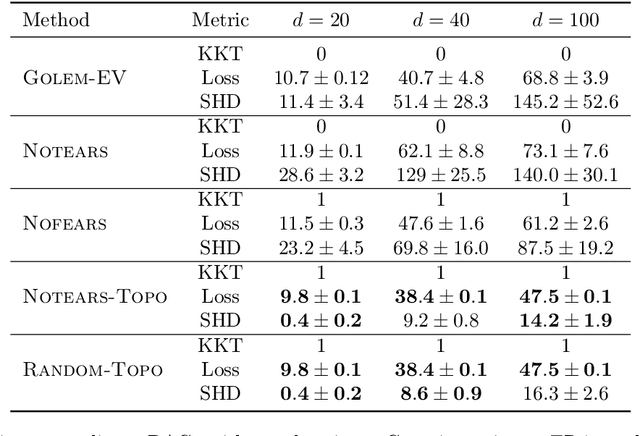
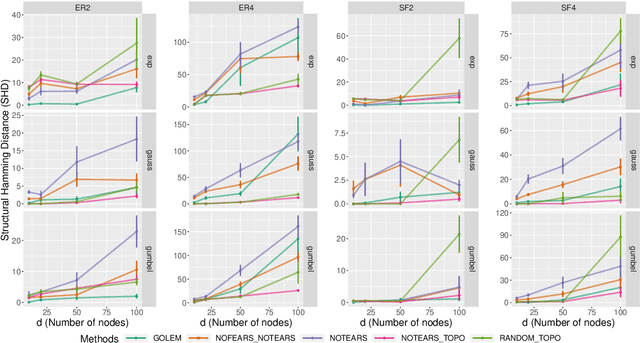
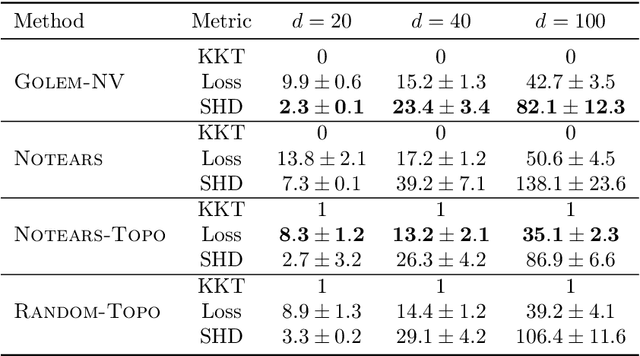
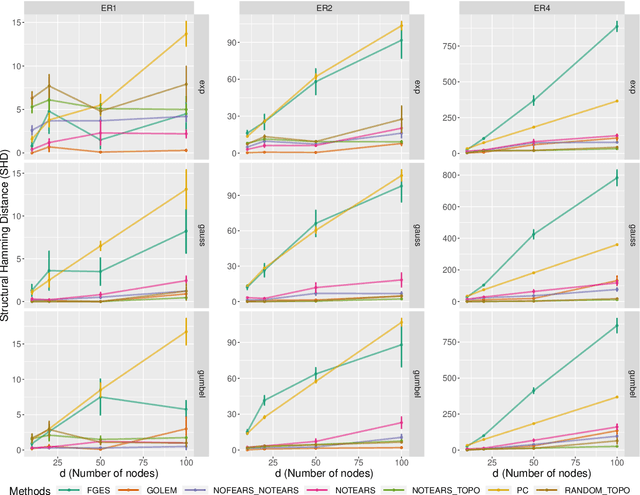
Recently, an intriguing class of non-convex optimization problems has emerged in the context of learning directed acyclic graphs (DAGs). These problems involve minimizing a given loss or score function, subject to a non-convex continuous constraint that penalizes the presence of cycles in a graph. In this work, we delve into the optimization challenges associated with this class of non-convex programs. To address these challenges, we propose a bi-level algorithm that leverages the non-convex constraint in a novel way. The outer level of the algorithm optimizes over topological orders by iteratively swapping pairs of nodes within the topological order of a DAG. A key innovation of our approach is the development of an effective method for generating a set of candidate swapping pairs for each iteration. At the inner level, given a topological order, we utilize off-the-shelf solvers that can handle linear constraints. The key advantage of our proposed algorithm is that it is guaranteed to find a local minimum or a KKT point under weaker conditions compared to previous work and finds solutions with lower scores. Extensive experiments demonstrate that our method outperforms state-of-the-art approaches in terms of achieving a better score. Additionally, our method can also be used as a post-processing algorithm to significantly improve the score of other algorithms. Code implementing the proposed method is available at https://github.com/duntrain/topo.
Learning Mixtures of Gaussians with Censored Data
May 06, 2023Wai Ming Tai, Bryon Aragam
We study the problem of learning mixtures of Gaussians with censored data. Statistical learning with censored data is a classical problem, with numerous practical applications, however, finite-sample guarantees for even simple latent variable models such as Gaussian mixtures are missing. Formally, we are given censored data from a mixture of univariate Gaussians $$\sum_{i=1}^k w_i \mathcal{N}(\mu_i,\sigma^2),$$ i.e. the sample is observed only if it lies inside a set $S$. The goal is to learn the weights $w_i$ and the means $\mu_i$. We propose an algorithm that takes only $\frac{1}{\varepsilon^{O(k)}}$ samples to estimate the weights $w_i$ and the means $\mu_i$ within $\varepsilon$ error.
DAGMA: Learning DAGs via M-matrices and a Log-Determinant Acyclicity Characterization
Sep 16, 2022Kevin Bello, Bryon Aragam, Pradeep Ravikumar
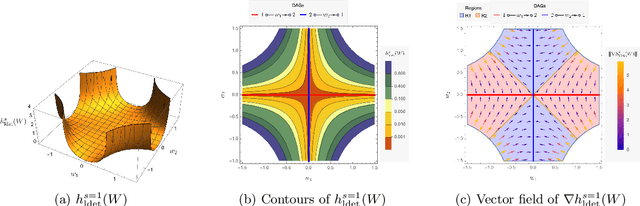
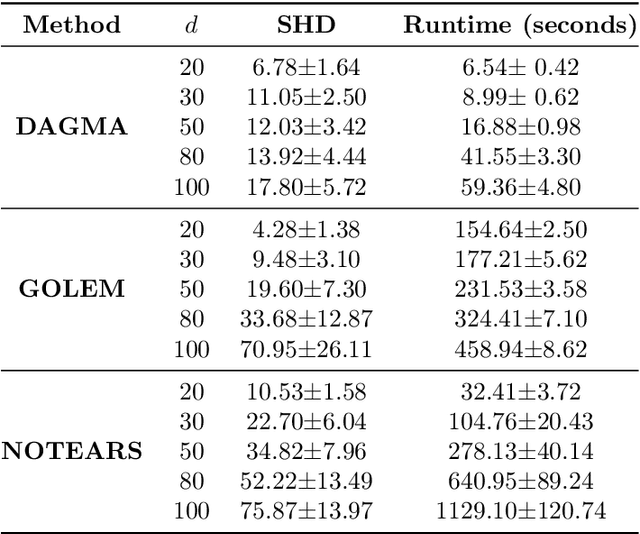
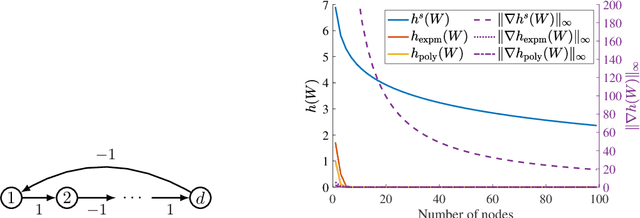
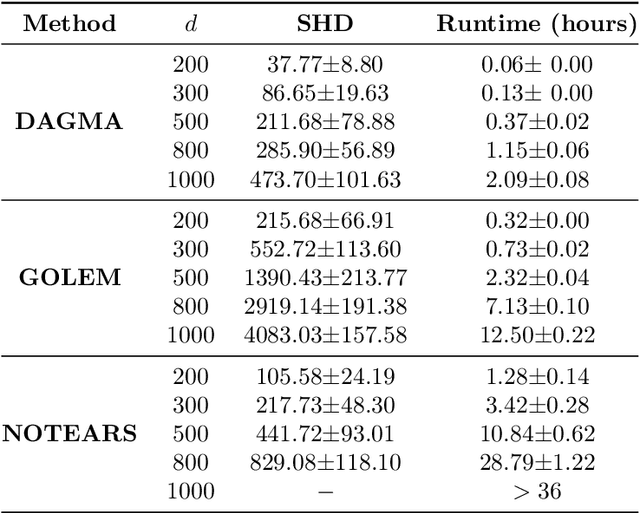
The combinatorial problem of learning directed acyclic graphs (DAGs) from data was recently framed as a purely continuous optimization problem by leveraging a differentiable acyclicity characterization of DAGs based on the trace of a matrix exponential function. Existing acyclicity characterizations are based on the idea that powers of an adjacency matrix contain information about walks and cycles. In this work, we propose a $\textit{fundamentally different}$ acyclicity characterization based on the log-determinant (log-det) function, which leverages the nilpotency property of DAGs. To deal with the inherent asymmetries of a DAG, we relate the domain of our log-det characterization to the set of $\textit{M-matrices}$, which is a key difference to the classical log-det function defined over the cone of positive definite matrices. Similar to acyclicity functions previously proposed, our characterization is also exact and differentiable. However, when compared to existing characterizations, our log-det function: (1) Is better at detecting large cycles; (2) Has better-behaved gradients; and (3) Its runtime is in practice about an order of magnitude faster. From the optimization side, we drop the typically used augmented Lagrangian scheme, and propose DAGMA ($\textit{Directed Acyclic Graphs via M-matrices for Acyclicity}$), a method that resembles the central path for barrier methods. Each point in the central path of DAGMA is a solution to an unconstrained problem regularized by our log-det function, then we show that at the limit of the central path the solution is guaranteed to be a DAG. Finally, we provide extensive experiments for $\textit{linear}$ and $\textit{nonlinear}$ SEMs, and show that our approach can reach large speed-ups and smaller structural Hamming distances against state-of-the-art methods.
Identifiability of deep generative models under mixture priors without auxiliary information
Jun 20, 2022Bohdan Kivva, Goutham Rajendran, Pradeep Ravikumar, Bryon Aragam
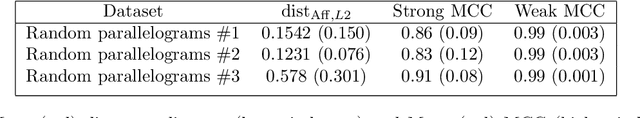

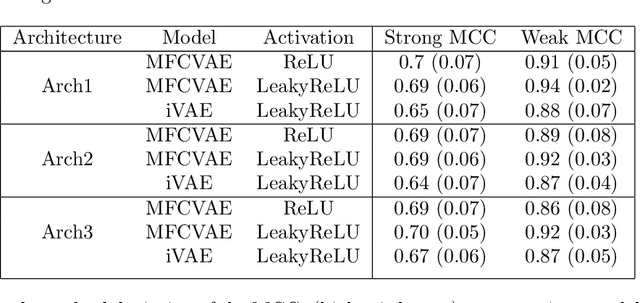
We prove identifiability of a broad class of deep latent variable models that (a) have universal approximation capabilities and (b) are the decoders of variational autoencoders that are commonly used in practice. Unlike existing work, our analysis does not require weak supervision, auxiliary information, or conditioning in the latent space. Recently, there has been a surge of works studying identifiability of such models. In these works, the main assumption is that along with the data, an auxiliary variable $u$ (also known as side information) is observed as well. At the same time, several works have empirically observed that this doesn't seem to be necessary in practice. In this work, we explain this behavior by showing that for a broad class of generative (i.e. unsupervised) models with universal approximation capabilities, the side information $u$ is not necessary: We prove identifiability of the entire generative model where we do not observe $u$ and only observe the data $x$. The models we consider are tightly connected with autoencoder architectures used in practice that leverage mixture priors in the latent space and ReLU/leaky-ReLU activations in the encoder. Our main result is an identifiability hierarchy that significantly generalizes previous work and exposes how different assumptions lead to different "strengths" of identifiability. For example, our weakest result establishes (unsupervised) identifiability up to an affine transformation, which already improves existing work. It's well known that these models have universal approximation capabilities and moreover, they have been extensively used in practice to learn representations of data.
A non-graphical representation of conditional independence via the neighbourhood lattice
Jun 12, 2022Arash A. Amini, Bryon Aragam, Qing Zhou
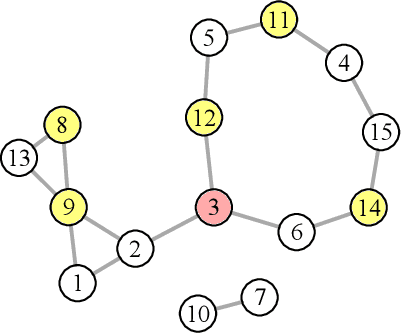
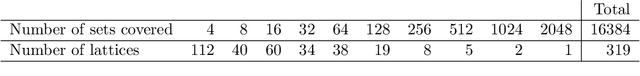
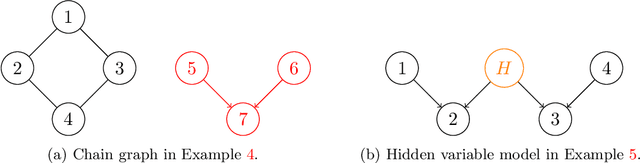
We introduce and study the neighbourhood lattice decomposition of a distribution, which is a compact, non-graphical representation of conditional independence that is valid in the absence of a faithful graphical representation. The idea is to view the set of neighbourhoods of a variable as a subset lattice, and partition this lattice into convex sublattices, each of which directly encodes a collection of conditional independence relations. We show that this decomposition exists in any compositional graphoid and can be computed efficiently and consistently in high-dimensions. {In particular, this gives a way to encode all of independence relations implied by a distribution that satisfies the composition axiom, which is strictly weaker than the faithfulness assumption that is typically assumed by graphical approaches.} We also discuss various special cases such as graphical models and projection lattices, each of which has intuitive interpretations. Along the way, we see how this problem is closely related to neighbourhood regression, which has been extensively studied in the context of graphical models and structural equations.
A super-polynomial lower bound for learning nonparametric mixtures
Mar 28, 2022Bryon Aragam, Wai Ming Tai
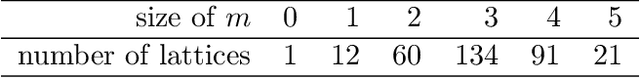
We study the problem of learning nonparametric distributions in a finite mixture, and establish a super-polynomial lower bound on the sample complexity of learning the component distributions in such models. Namely, we are given i.i.d. samples from $f$ where $$ f=\sum_{i=1}^k w_i f_i, \quad\sum_{i=1}^k w_i=1, \quad w_i>0 $$ and we are interested in learning each component $f_i$. Without any assumptions on $f_i$, this problem is ill-posed. In order to identify the components $f_i$, we assume that each $f_i$ can be written as a convolution of a Gaussian and a compactly supported density $\nu_i$ with $\text{supp}(\nu_i)\cap \text{supp}(\nu_j)=\emptyset$. Our main result shows that $\Omega((\frac{1}{\varepsilon})^{C\log\log \frac{1}{\varepsilon}})$ samples are required for estimating each $f_i$. The proof relies on a fast rate for approximation with Gaussians, which may be of independent interest. This result has important implications for the hardness of learning more general nonparametric latent variable models that arise in machine learning applications.
Optimal estimation of Gaussian DAG models
Jan 25, 2022Ming Gao, Wai Ming Tai, Bryon Aragam
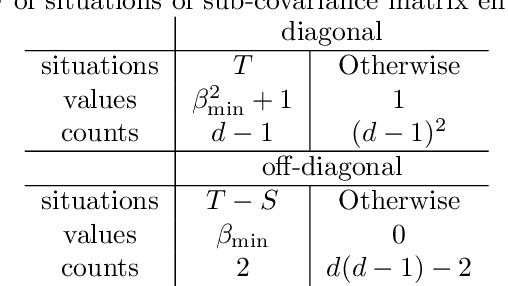
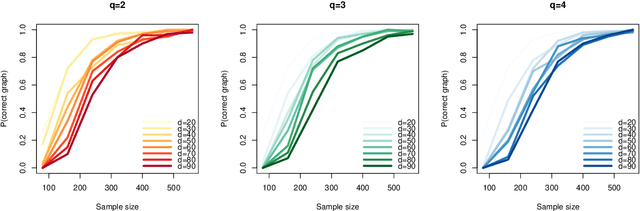
We study the optimal sample complexity of learning a Gaussian directed acyclic graph (DAG) from observational data. Our main result establishes the minimax optimal sample complexity for learning the structure of a linear Gaussian DAG model with equal variances to be $n\asymp q\log(d/q)$, where $q$ is the maximum number of parents and $d$ is the number of nodes. We further make comparisons with the classical problem of learning (undirected) Gaussian graphical models, showing that under the equal variance assumption, these two problems share the same optimal sample complexity. In other words, at least for Gaussian models with equal error variances, learning a directed graphical model is not more difficult than learning an undirected graphical model. Our results also extend to more general identification assumptions as well as subgaussian errors.
Tradeoffs of Linear Mixed Models in Genome-wide Association Studies
Nov 05, 2021Haohan Wang, Bryon Aragam, Eric Xing
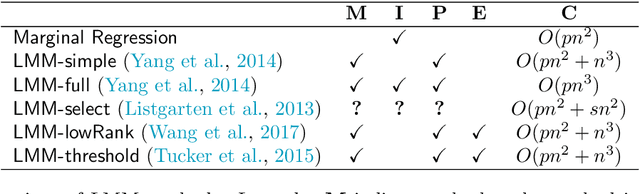
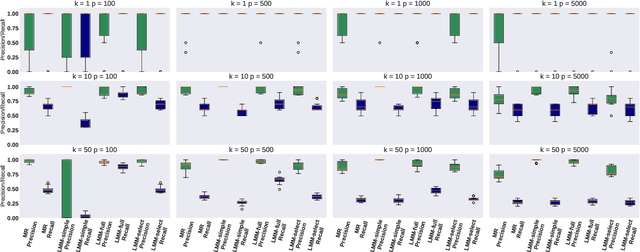
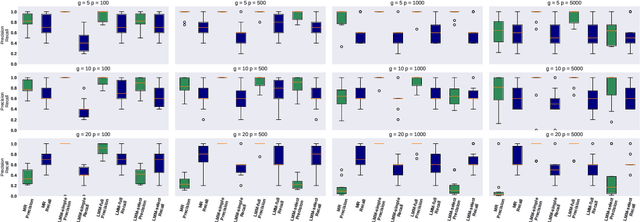
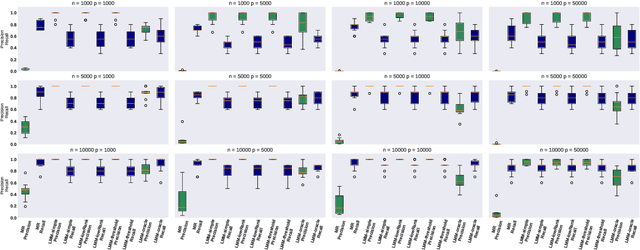
Motivated by empirical arguments that are well-known from the genome-wide association studies (GWAS) literature, we study the statistical properties of linear mixed models (LMMs) applied to GWAS. First, we study the sensitivity of LMMs to the inclusion of a candidate SNP in the kinship matrix, which is often done in practice to speed up computations. Our results shed light on the size of the error incurred by including a candidate SNP, providing a justification to this technique in order to trade-off velocity against veracity. Second, we investigate how mixed models can correct confounders in GWAS, which is widely accepted as an advantage of LMMs over traditional methods. We consider two sources of confounding factors, population stratification and environmental confounding factors, and study how different methods that are commonly used in practice trade-off these two confounding factors differently.
NOTMAD: Estimating Bayesian Networks with Sample-Specific Structures and Parameters
Nov 01, 2021Ben Lengerich, Caleb Ellington, Bryon Aragam, Eric P. Xing, Manolis Kellis
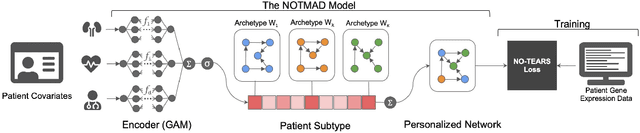


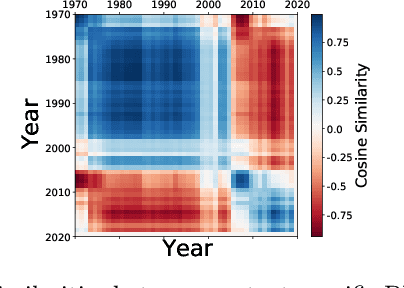
Context-specific Bayesian networks (i.e. directed acyclic graphs, DAGs) identify context-dependent relationships between variables, but the non-convexity induced by the acyclicity requirement makes it difficult to share information between context-specific estimators (e.g. with graph generator functions). For this reason, existing methods for inferring context-specific Bayesian networks have favored breaking datasets into subsamples, limiting statistical power and resolution, and preventing the use of multidimensional and latent contexts. To overcome this challenge, we propose NOTEARS-optimized Mixtures of Archetypal DAGs (NOTMAD). NOTMAD models context-specific Bayesian networks as the output of a function which learns to mix archetypal networks according to sample context. The archetypal networks are estimated jointly with the context-specific networks and do not require any prior knowledge. We encode the acyclicity constraint as a smooth regularization loss which is back-propagated to the mixing function; in this way, NOTMAD shares information between context-specific acyclic graphs, enabling the estimation of Bayesian network structures and parameters at even single-sample resolution. We demonstrate the utility of NOTMAD and sample-specific network inference through analysis and experiments, including patient-specific gene expression networks which correspond to morphological variation in cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge