Boyun Zheng
MedSAM-Agent: Empowering Interactive Medical Image Segmentation with Multi-turn Agentic Reinforcement Learning
Feb 03, 2026Abstract:Medical image segmentation is evolving from task-specific models toward generalizable frameworks. Recent research leverages Multi-modal Large Language Models (MLLMs) as autonomous agents, employing reinforcement learning with verifiable reward (RLVR) to orchestrate specialized tools like the Segment Anything Model (SAM). However, these approaches often rely on single-turn, rigid interaction strategies and lack process-level supervision during training, which hinders their ability to fully exploit the dynamic potential of interactive tools and leads to redundant actions. To bridge this gap, we propose MedSAM-Agent, a framework that reformulates interactive segmentation as a multi-step autonomous decision-making process. First, we introduce a hybrid prompting strategy for expert-curated trajectory generation, enabling the model to internalize human-like decision heuristics and adaptive refinement strategies. Furthermore, we develop a two-stage training pipeline that integrates multi-turn, end-to-end outcome verification with a clinical-fidelity process reward design to promote interaction parsimony and decision efficiency. Extensive experiments across 6 medical modalities and 21 datasets demonstrate that MedSAM-Agent achieves state-of-the-art performance, effectively unifying autonomous medical reasoning with robust, iterative optimization. Code is available \href{https://github.com/CUHK-AIM-Group/MedSAM-Agent}{here}.
TumorGen: Boundary-Aware Tumor-Mask Synthesis with Rectified Flow Matching
May 30, 2025



Abstract:Tumor data synthesis offers a promising solution to the shortage of annotated medical datasets. However, current approaches either limit tumor diversity by using predefined masks or employ computationally expensive two-stage processes with multiple denoising steps, causing computational inefficiency. Additionally, these methods typically rely on binary masks that fail to capture the gradual transitions characteristic of tumor boundaries. We present TumorGen, a novel Boundary-Aware Tumor-Mask Synthesis with Rectified Flow Matching for efficient 3D tumor synthesis with three key components: a Boundary-Aware Pseudo Mask Generation module that replaces strict binary masks with flexible bounding boxes; a Spatial-Constraint Vector Field Estimator that simultaneously synthesizes tumor latents and masks using rectified flow matching to ensure computational efficiency; and a VAE-guided mask refiner that enhances boundary realism. TumorGen significantly improves computational efficiency by requiring fewer sampling steps while maintaining pathological accuracy through coarse and fine-grained spatial constraints. Experimental results demonstrate TumorGen's superior performance over existing tumor synthesis methods in both efficiency and realism, offering a valuable contribution to AI-driven cancer diagnostics.
A Comprehensive Evaluation of Multi-Modal Large Language Models for Endoscopy Analysis
May 29, 2025
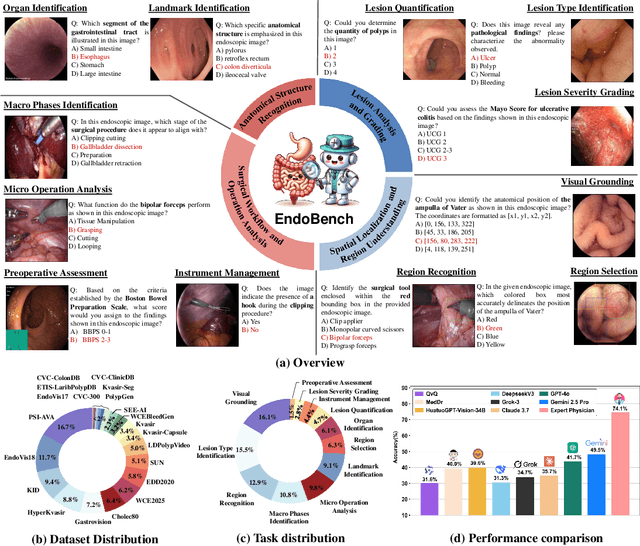


Abstract:Endoscopic procedures are essential for diagnosing and treating internal diseases, and multi-modal large language models (MLLMs) are increasingly applied to assist in endoscopy analysis. However, current benchmarks are limited, as they typically cover specific endoscopic scenarios and a small set of clinical tasks, failing to capture the real-world diversity of endoscopic scenarios and the full range of skills needed in clinical workflows. To address these issues, we introduce EndoBench, the first comprehensive benchmark specifically designed to assess MLLMs across the full spectrum of endoscopic practice with multi-dimensional capacities. EndoBench encompasses 4 distinct endoscopic scenarios, 12 specialized clinical tasks with 12 secondary subtasks, and 5 levels of visual prompting granularities, resulting in 6,832 rigorously validated VQA pairs from 21 diverse datasets. Our multi-dimensional evaluation framework mirrors the clinical workflow--spanning anatomical recognition, lesion analysis, spatial localization, and surgical operations--to holistically gauge the perceptual and diagnostic abilities of MLLMs in realistic scenarios. We benchmark 23 state-of-the-art models, including general-purpose, medical-specialized, and proprietary MLLMs, and establish human clinician performance as a reference standard. Our extensive experiments reveal: (1) proprietary MLLMs outperform open-source and medical-specialized models overall, but still trail human experts; (2) medical-domain supervised fine-tuning substantially boosts task-specific accuracy; and (3) model performance remains sensitive to prompt format and clinical task complexity. EndoBench establishes a new standard for evaluating and advancing MLLMs in endoscopy, highlighting both progress and persistent gaps between current models and expert clinical reasoning. We publicly release our benchmark and code.
Pathological Semantics-Preserving Learning for H&E-to-IHC Virtual Staining
Jul 04, 2024



Abstract:Conventional hematoxylin-eosin (H&E) staining is limited to revealing cell morphology and distribution, whereas immunohistochemical (IHC) staining provides precise and specific visualization of protein activation at the molecular level. Virtual staining technology has emerged as a solution for highly efficient IHC examination, which directly transforms H&E-stained images to IHC-stained images. However, virtual staining is challenged by the insufficient mining of pathological semantics and the spatial misalignment of pathological semantics. To address these issues, we propose the Pathological Semantics-Preserving Learning method for Virtual Staining (PSPStain), which directly incorporates the molecular-level semantic information and enhances semantics interaction despite any spatial inconsistency. Specifically, PSPStain comprises two novel learning strategies: 1) Protein-Aware Learning Strategy (PALS) with Focal Optical Density (FOD) map maintains the coherence of protein expression level, which represents molecular-level semantic information; 2) Prototype-Consistent Learning Strategy (PCLS), which enhances cross-image semantic interaction by prototypical consistency learning. We evaluate PSPStain on two public datasets using five metrics: three clinically relevant metrics and two for image quality. Extensive experiments indicate that PSPStain outperforms current state-of-the-art H&E-to-IHC virtual staining methods and demonstrates a high pathological correlation between the staging of real and virtual stains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge