Bin Yu
Towards Robust Waveform-Based Acoustic Models
Oct 16, 2021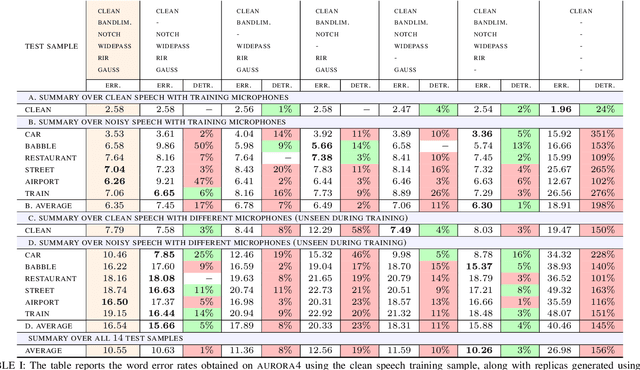
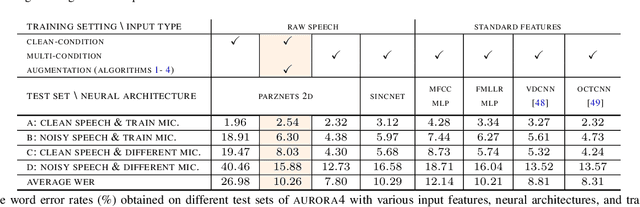
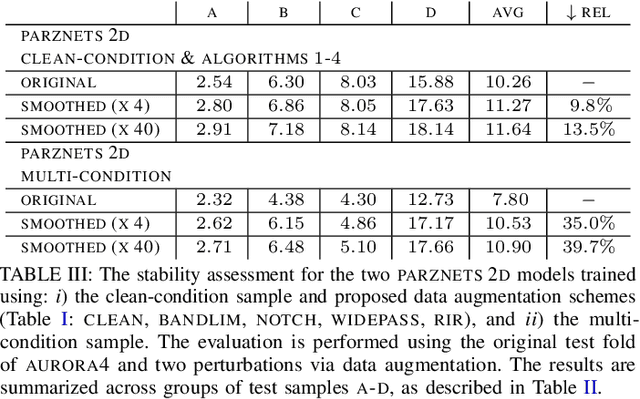
Abstract:We propose an approach for learning robust acoustic models in adverse environments, characterized by a significant mismatch between training and test conditions. This problem is of paramount importance for the deployment of speech recognition systems that need to perform well in unseen environments. Our approach is an instance of vicinal risk minimization, which aims to improve risk estimates during training by replacing the delta functions that define the empirical density over the input space with an approximation of the marginal population density in the vicinity of the training samples. More specifically, we assume that local neighborhoods centered at training samples can be approximated using a mixture of Gaussians, and demonstrate theoretically that this can incorporate robust inductive bias into the learning process. We characterize the individual mixture components implicitly via data augmentation schemes, designed to address common sources of spurious correlations in acoustic models. To avoid potential confounding effects on robustness due to information loss, which has been associated with standard feature extraction techniques (e.g., FBANK and MFCC features), we focus our evaluation on the waveform-based setting. Our empirical results show that the proposed approach can generalize to unseen noise conditions, with 150% relative improvement in out-of-distribution generalization compared to training using the standard risk minimization principle. Moreover, the results demonstrate competitive performance relative to models learned using a training sample designed to match the acoustic conditions characteristic of test utterances (i.e., optimal vicinal densities).
Interpreting and improving deep-learning models with reality checks
Aug 19, 2021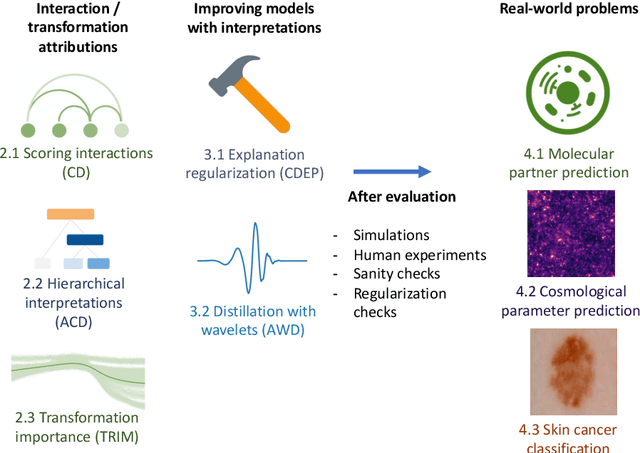

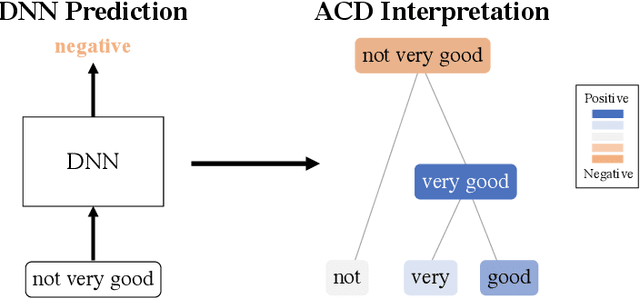
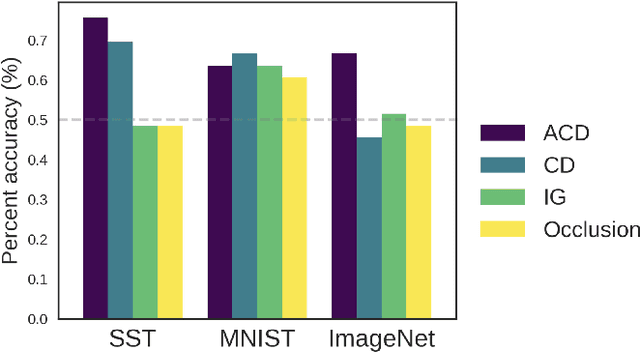
Abstract:Recent deep-learning models have achieved impressive predictive performance by learning complex functions of many variables, often at the cost of interpretability. This chapter covers recent work aiming to interpret models by attributing importance to features and feature groups for a single prediction. Importantly, the proposed attributions assign importance to interactions between features, in addition to features in isolation. These attributions are shown to yield insights across real-world domains, including bio-imaging, cosmology image and natural-language processing. We then show how these attributions can be used to directly improve the generalization of a neural network or to distill it into a simple model. Throughout the chapter, we emphasize the use of reality checks to scrutinize the proposed interpretation techniques.
Adaptive wavelet distillation from neural networks through interpretations
Jul 19, 2021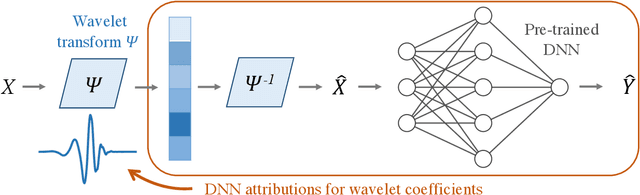
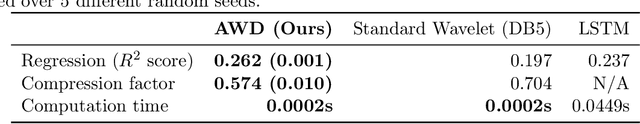
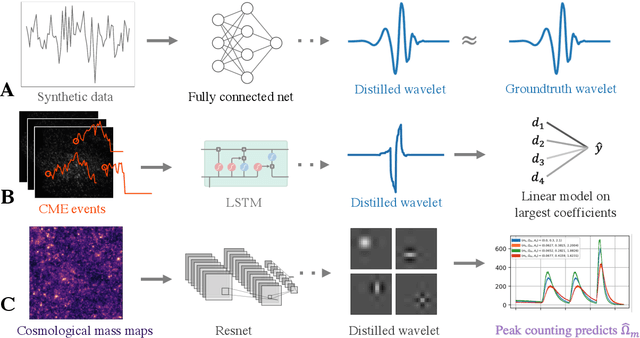

Abstract:Recent deep-learning models have achieved impressive prediction performance, but often sacrifice interpretability and computational efficiency. Interpretability is crucial in many disciplines, such as science and medicine, where models must be carefully vetted or where interpretation is the goal itself. Moreover, interpretable models are concise and often yield computational efficiency. Here, we propose adaptive wavelet distillation (AWD), a method which aims to distill information from a trained neural network into a wavelet transform. Specifically, AWD penalizes feature attributions of a neural network in the wavelet domain to learn an effective multi-resolution wavelet transform. The resulting model is highly predictive, concise, computationally efficient, and has properties (such as a multi-scale structure) which make it easy to interpret. In close collaboration with domain experts, we showcase how AWD addresses challenges in two real-world settings: cosmological parameter inference and molecular-partner prediction. In both cases, AWD yields a scientifically interpretable and concise model which gives predictive performance better than state-of-the-art neural networks. Moreover, AWD identifies predictive features that are scientifically meaningful in the context of respective domains. All code and models are released in a full-fledged package available on Github (https://github.com/Yu-Group/adaptive-wavelets).
Enriched Annotations for Tumor Attribute Classification from Pathology Reports with Limited Labeled Data
Dec 15, 2020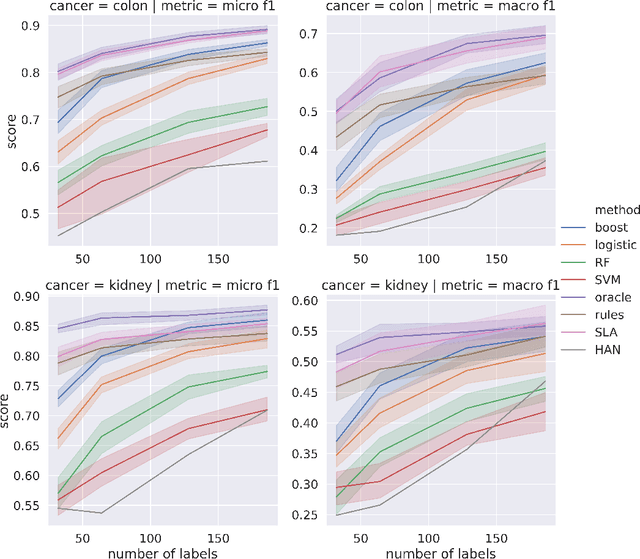
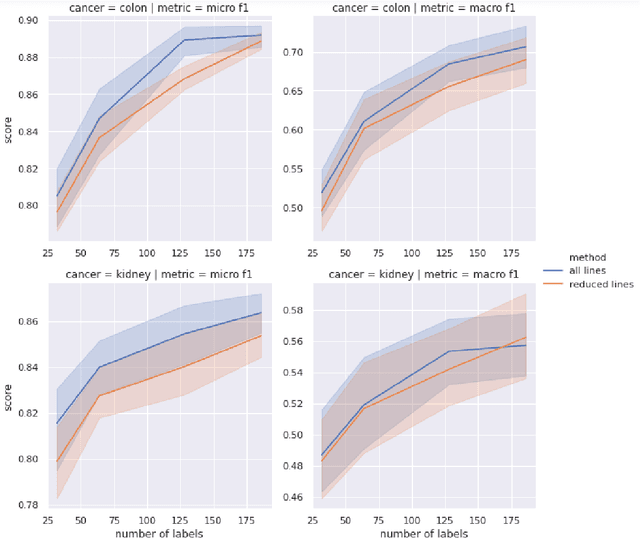
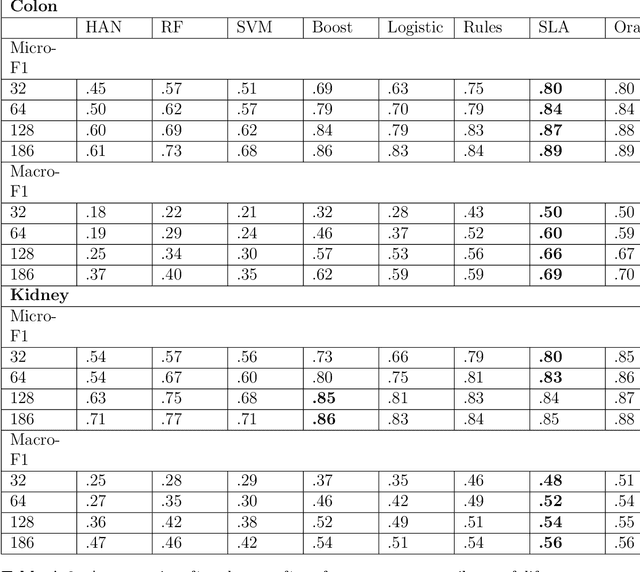
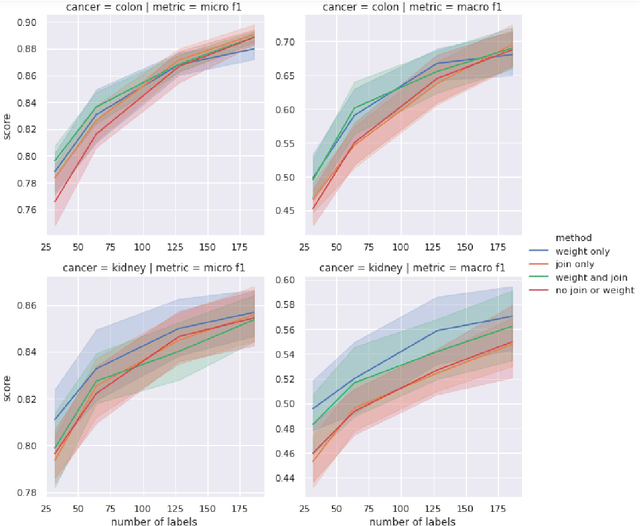
Abstract:Precision medicine has the potential to revolutionize healthcare, but much of the data for patients is locked away in unstructured free-text, limiting research and delivery of effective personalized treatments. Generating large annotated datasets for information extraction from clinical notes is often challenging and expensive due to the high level of expertise needed for high quality annotations. To enable natural language processing for small dataset sizes, we develop a novel enriched hierarchical annotation scheme and algorithm, Supervised Line Attention (SLA), and apply this algorithm to predicting categorical tumor attributes from kidney and colon cancer pathology reports from the University of California San Francisco (UCSF). Whereas previous work only annotated document level labels, we in addition ask the annotators to enrich the traditional label by asking them to also highlight the relevant line or potentially lines for the final label, which leads to a 20% increase of annotation time required per document. With the enriched annotations, we develop a simple and interpretable machine learning algorithm that first predicts the relevant lines in the document and then predicts the tumor attribute. Our results show across the small dataset sizes of 32, 64, 128, and 186 labeled documents per cancer, SLA only requires half the number of labeled documents as state-of-the-art methods to achieve similar or better micro-f1 and macro-f1 scores for the vast majority of comparisons that we made. Accounting for the increased annotation time, this leads to a 40% reduction in total annotation time over the state of the art.
Stable discovery of interpretable subgroups via calibration in causal studies
Sep 29, 2020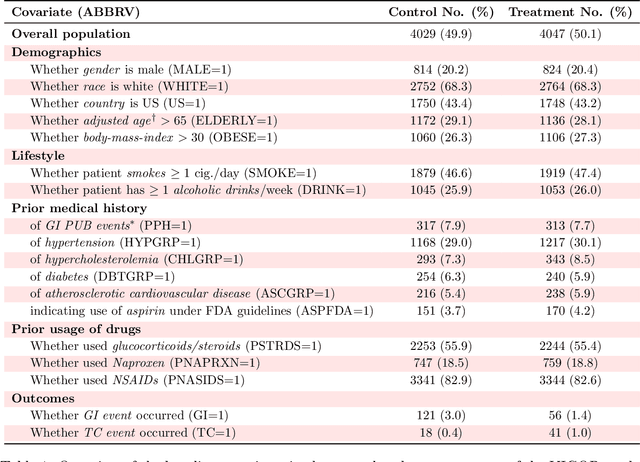
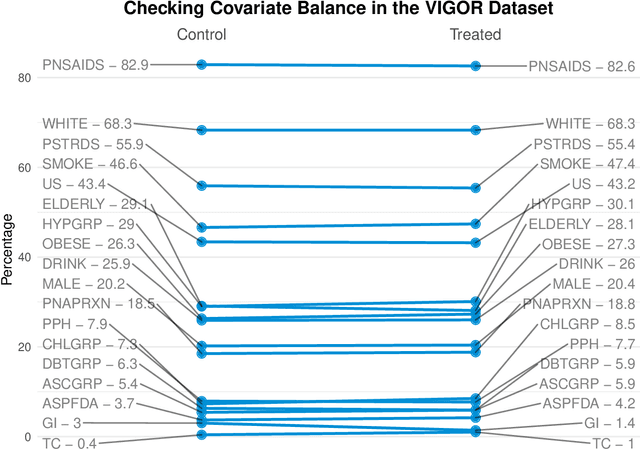
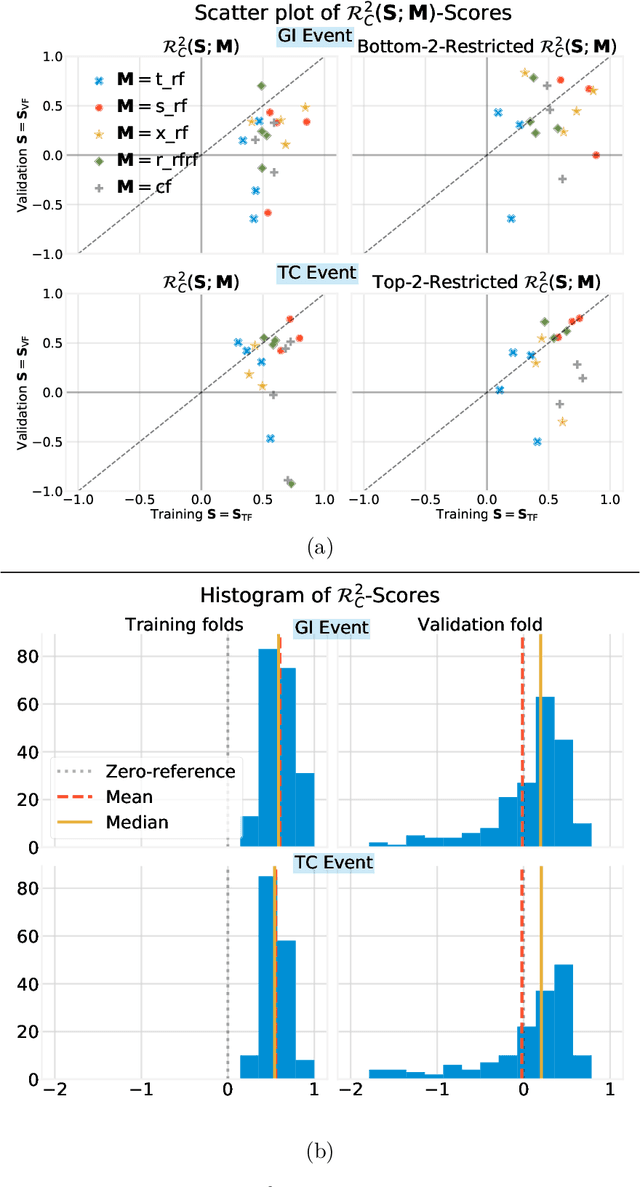
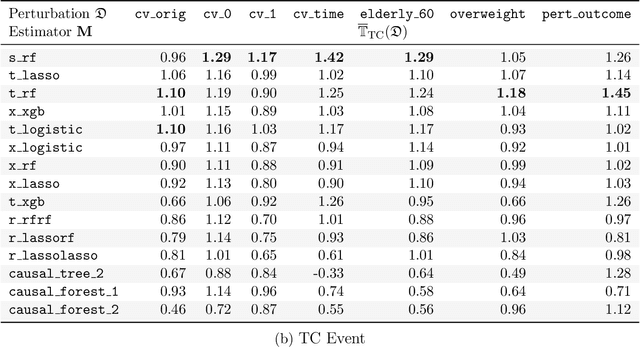
Abstract:Building on Yu and Kumbier's PCS framework and for randomized experiments, we introduce a novel methodology for Stable Discovery of Interpretable Subgroups via Calibration (StaDISC), with large heterogeneous treatment effects. StaDISC was developed during our re-analysis of the 1999-2000 VIGOR study, an 8076 patient randomized controlled trial (RCT), that compared the risk of adverse events from a then newly approved drug, Rofecoxib (Vioxx), to that from an older drug Naproxen. Vioxx was found to, on average and in comparison to Naproxen, reduce the risk of gastrointestinal (GI) events but increase the risk of thrombotic cardiovascular (CVT) events. Applying StaDISC, we fit 18 popular conditional average treatment effect (CATE) estimators for both outcomes and use calibration to demonstrate their poor global performance. However, they are locally well-calibrated and stable, enabling the identification of patient groups with larger than (estimated) average treatment effects. In fact, StaDISC discovers three clinically interpretable subgroups each for the GI outcome (totaling 29.4% of the study size) and the CVT outcome (totaling 11.0%). Complementary analyses of the found subgroups using the 2001-2004 APPROVe study, a separate independently conducted RCT with 2587 patients, provides further supporting evidence for the promise of StaDISC.
Revisiting complexity and the bias-variance tradeoff
Jun 17, 2020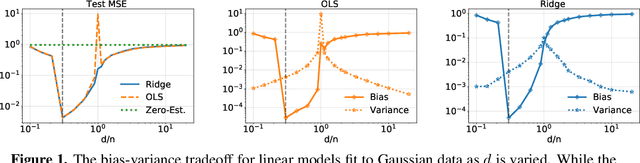
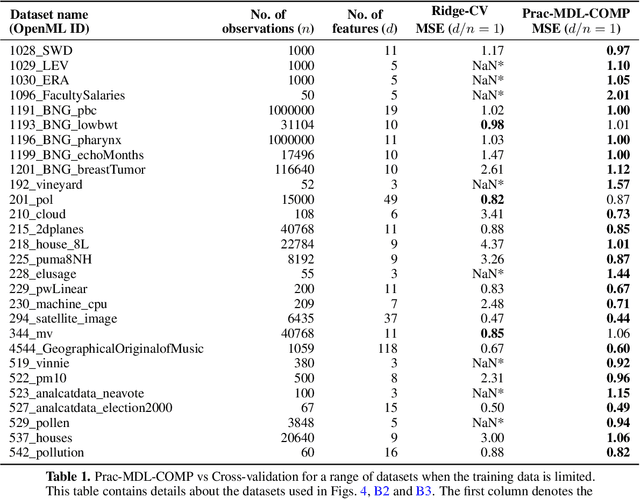
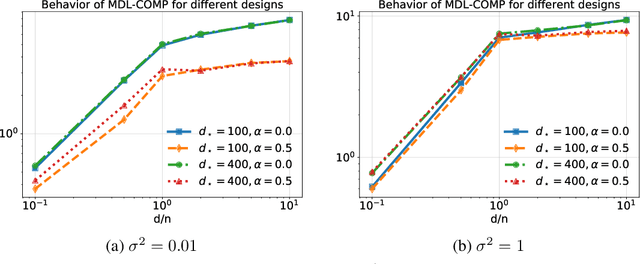
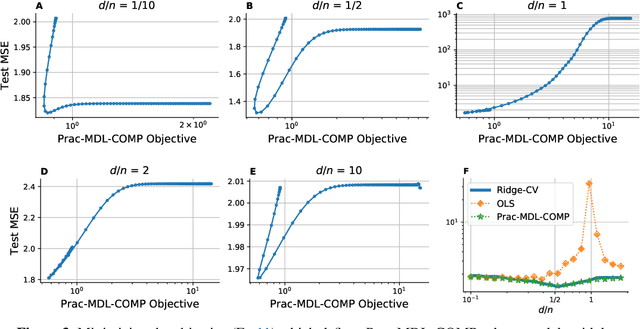
Abstract:The recent success of high-dimensional models, such as deep neural networks (DNNs), has led many to question the validity of the bias-variance tradeoff principle in high dimensions. We reexamine it with respect to two key choices: the model class and the complexity measure. We argue that failing to suitably specify either one can falsely suggest that the tradeoff does not hold. This observation motivates us to seek a valid complexity measure, defined with respect to a reasonably good class of models. Building on Rissanen's principle of minimum description length (MDL), we propose a novel MDL-based complexity (MDL-COMP). We focus on the context of linear models, which have been recently used as a stylized tractable approximation to DNNs in high-dimensions. MDL-COMP is defined via an optimality criterion over the encodings induced by a good Ridge estimator class. We derive closed-form expressions for MDL-COMP and show that for a dataset with $n$ observations and $d$ parameters it is \emph{not always} equal to $d/n$, and is a function of the singular values of the design matrix and the signal-to-noise ratio. For random Gaussian design, we find that while MDL-COMP scales linearly with $d$ in low-dimensions ($d<n$), for high-dimensions ($d>n$) the scaling is exponentially smaller, scaling as $\log d$. We hope that such a slow growth of complexity in high-dimensions can help shed light on the good generalization performance of several well-tuned high-dimensional models. Moreover, via an array of simulations and real-data experiments, we show that a data-driven Prac-MDL-COMP can inform hyper-parameter tuning for ridge regression in limited data settings, sometimes improving upon cross-validation.
Instability, Computational Efficiency and Statistical Accuracy
May 22, 2020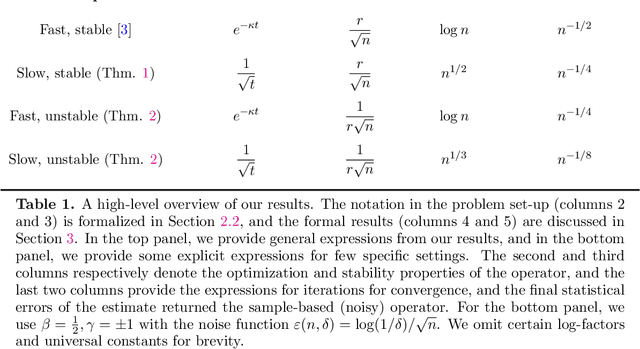
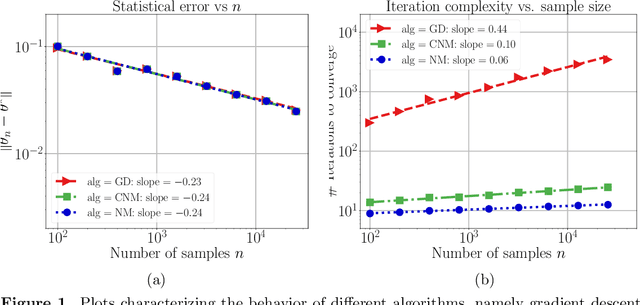
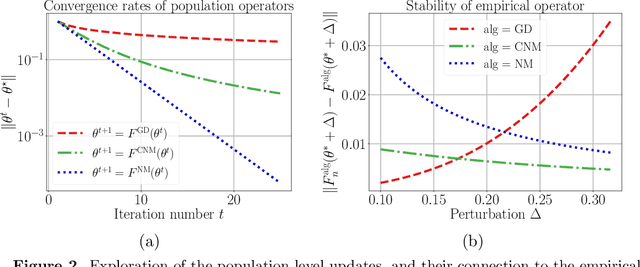
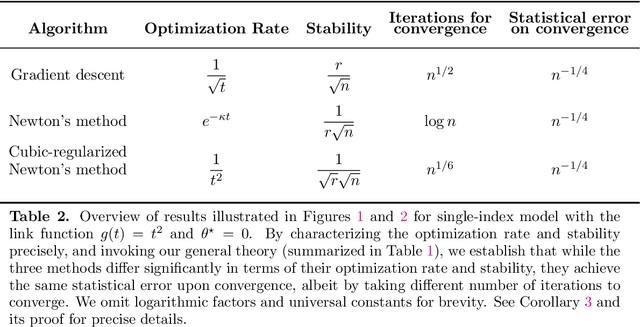
Abstract:Many statistical estimators are defined as the fixed point of a data-dependent operator, with estimators based on minimizing a cost function being an important special case. The limiting performance of such estimators depends on the properties of the population-level operator in the idealized limit of infinitely many samples. We develop a general framework that yields bounds on statistical accuracy based on the interplay between the deterministic convergence rate of the algorithm at the population level, and its degree of (in)stability when applied to an empirical object based on $n$ samples. Using this framework, we analyze both stable forms of gradient descent and some higher-order and unstable algorithms, including Newton's method and its cubic-regularized variant, as well as the EM algorithm. We provide applications of our general results to several concrete classes of models, including Gaussian mixture estimation, single-index models, and informative non-response models. We exhibit cases in which an unstable algorithm can achieve the same statistical accuracy as a stable algorithm in exponentially fewer steps---namely, with the number of iterations being reduced from polynomial to logarithmic in sample size $n$.
Curating a COVID-19 data repository and forecasting county-level death counts in the United States
May 16, 2020
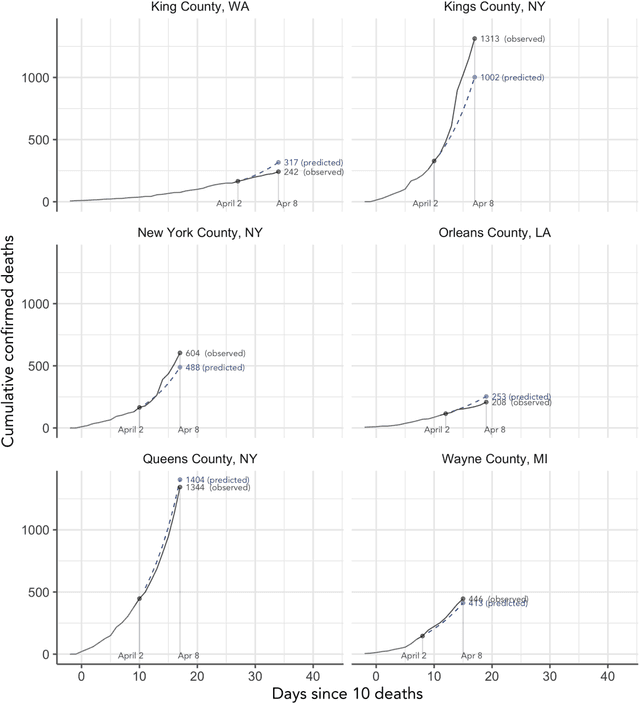
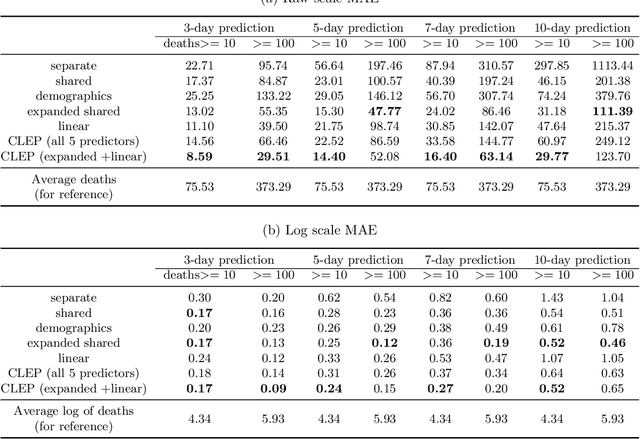
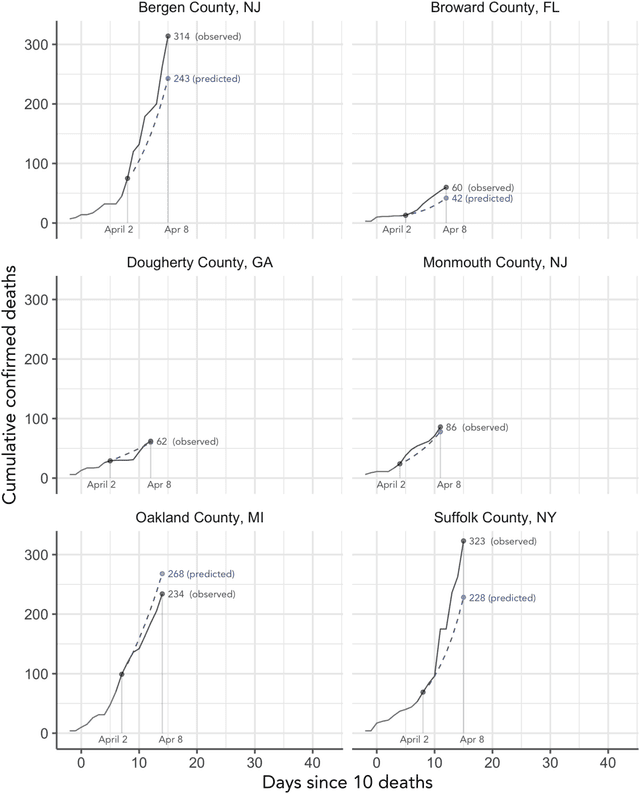
Abstract:As the COVID-19 outbreak continues to evolve, accurate forecasting continues to play an extremely important role in informing policy decisions. In this paper, we collate a large data repository containing COVID-19 information from a range of different sources. We use this data to develop several predictors and prediction intervals for forecasting the short-term (e.g., over the next week) trajectory of COVID-19-related recorded deaths at the county-level in the United States. Specifically, using data from January 22, 2020, to May 10, 2020, we produce several different predictors and combine their forecasts using ensembling techniques, resulting in an ensemble we refer to as Combined Linear and Exponential Predictors (CLEP). Our individual predictors include county-specific exponential and linear predictors, an exponential predictor that pools data together across counties, and a demographics-based exponential predictor. In addition, we use the largest prediction errors in the past five days to assess the uncertainty of our death predictions, resulting in prediction intervals that we refer to as Maximum (absolute) Error Prediction Intervals (MEPI). We show that MEPI is an effective method in practice with a 94.5\% coverage rate when averaged across counties. Our forecasts are already being used by the non-profit organization, Response4Life, to determine the medical supply need for individual hospitals and have directly contributed to the distribution of medical supplies across the country. We hope that our forecasts and data repository can help guide necessary county-specific decision-making and help counties prepare for their continued fight against COVID-19. All collected data, modeling code, forecasts, and visualizations are updated daily and available at \url{https://github.com/Yu-Group/covid19-severity-prediction}.
Transformation Importance with Applications to Cosmology
Mar 04, 2020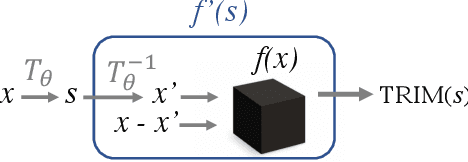
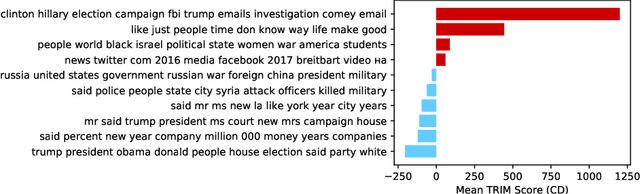
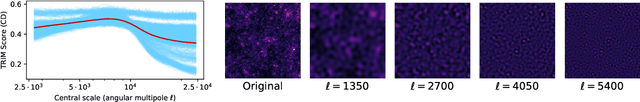
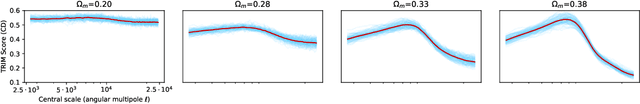
Abstract:Machine learning lies at the heart of new possibilities for scientific discovery, knowledge generation, and artificial intelligence. Its potential benefits to these fields requires going beyond predictive accuracy and focusing on interpretability. In particular, many scientific problems require interpretations in a domain-specific interpretable feature space (e.g. the frequency domain) whereas attributions to the raw features (e.g. the pixel space) may be unintelligible or even misleading. To address this challenge, we propose TRIM (TRansformation IMportance), a novel approach which attributes importances to features in a transformed space and can be applied post-hoc to a fully trained model. TRIM is motivated by a cosmological parameter estimation problem using deep neural networks (DNNs) on simulated data, but it is generally applicable across domains/models and can be combined with any local interpretation method. In our cosmology example, combining TRIM with contextual decomposition shows promising results for identifying which frequencies a DNN uses, helping cosmologists to understand and validate that the model learns appropriate physical features rather than simulation artifacts.
Interpretations are useful: penalizing explanations to align neural networks with prior knowledge
Oct 01, 2019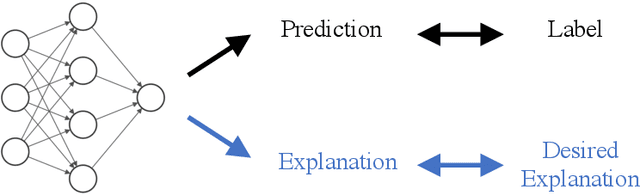

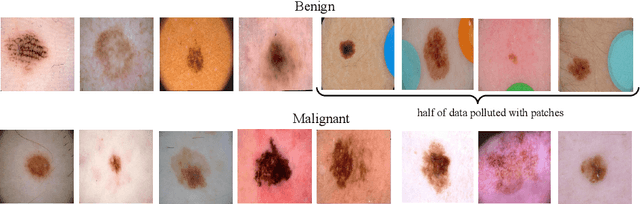
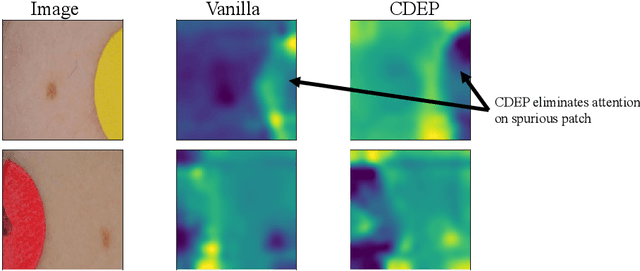
Abstract:For an explanation of a deep learning model to be effective, it must provide both insight into a model and suggest a corresponding action in order to achieve some objective. Too often, the litany of proposed explainable deep learning methods stop at the first step, providing practitioners with insight into a model, but no way to act on it. In this paper, we propose contextual decomposition explanation penalization (CDEP), a method which enables practitioners to leverage existing explanation methods in order to increase the predictive accuracy of deep learning models. In particular, when shown that a model has incorrectly assigned importance to some features, CDEP enables practitioners to correct these errors by directly regularizing the provided explanations. Using explanations provided by contextual decomposition (CD) (Murdoch et al., 2018), we demonstrate the ability of our method to increase performance on an array of toy and real datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge