Andrew P. King
King's College London
A Deep Learning-based Integrated Framework for Quality-aware Undersampled Cine Cardiac MRI Reconstruction and Analysis
May 02, 2022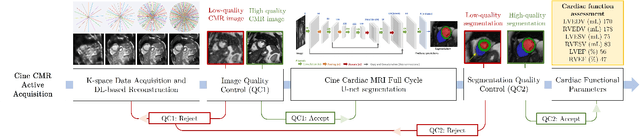
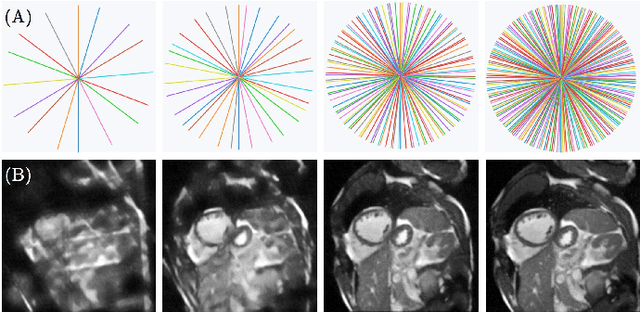
Abstract:Cine cardiac magnetic resonance (CMR) imaging is considered the gold standard for cardiac function evaluation. However, cine CMR acquisition is inherently slow and in recent decades considerable effort has been put into accelerating scan times without compromising image quality or the accuracy of derived results. In this paper, we present a fully-automated, quality-controlled integrated framework for reconstruction, segmentation and downstream analysis of undersampled cine CMR data. The framework enables active acquisition of radial k-space data, in which acquisition can be stopped as soon as acquired data are sufficient to produce high quality reconstructions and segmentations. This results in reduced scan times and automated analysis, enabling robust and accurate estimation of functional biomarkers. To demonstrate the feasibility of the proposed approach, we perform realistic simulations of radial k-space acquisitions on a dataset of subjects from the UK Biobank and present results on in-vivo cine CMR k-space data collected from healthy subjects. The results demonstrate that our method can produce quality-controlled images in a mean scan time reduced from 12 to 4 seconds per slice, and that image quality is sufficient to allow clinically relevant parameters to be automatically estimated to within 5% mean absolute difference.
AI-enabled Assessment of Cardiac Systolic and Diastolic Function from Echocardiography
Mar 21, 2022
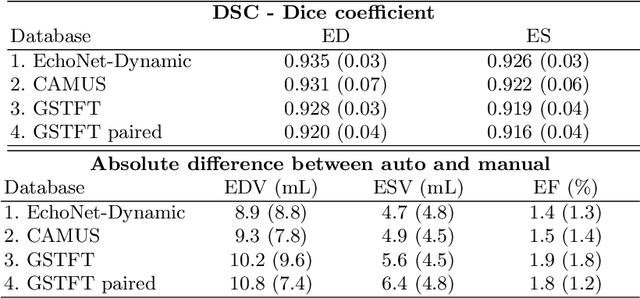
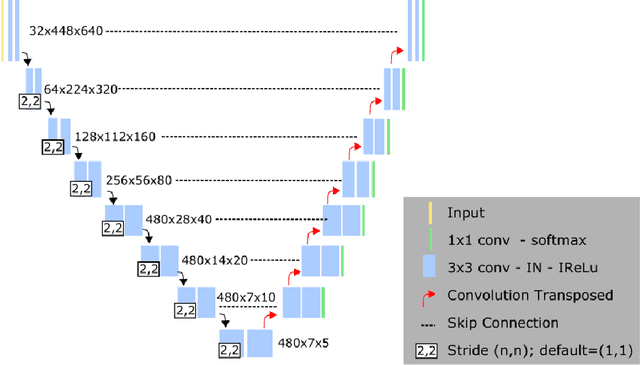
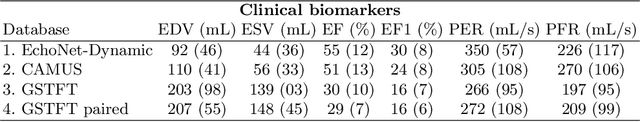
Abstract:Left ventricular (LV) function is an important factor in terms of patient management, outcome, and long-term survival of patients with heart disease. The most recently published clinical guidelines for heart failure recognise that over reliance on only one measure of cardiac function (LV ejection fraction) as a diagnostic and treatment stratification biomarker is suboptimal. Recent advances in AI-based echocardiography analysis have shown excellent results on automated estimation of LV volumes and LV ejection fraction. However, from time-varying 2-D echocardiography acquisition, a richer description of cardiac function can be obtained by estimating functional biomarkers from the complete cardiac cycle. In this work we propose for the first time an AI approach for deriving advanced biomarkers of systolic and diastolic LV function from 2-D echocardiography based on segmentations of the full cardiac cycle. These biomarkers will allow clinicians to obtain a much richer picture of the heart in health and disease. The AI model is based on the 'nn-Unet' framework and was trained and tested using four different databases. Results show excellent agreement between manual and automated analysis and showcase the potential of the advanced systolic and diastolic biomarkers for patient stratification. Finally, for a subset of 50 cases, we perform a correlation analysis between clinical biomarkers derived from echocardiography and CMR and we show excellent agreement between the two modalities.
The Impact of Domain Shift on Left and Right Ventricle Segmentation in Short Axis Cardiac MR Images
Sep 22, 2021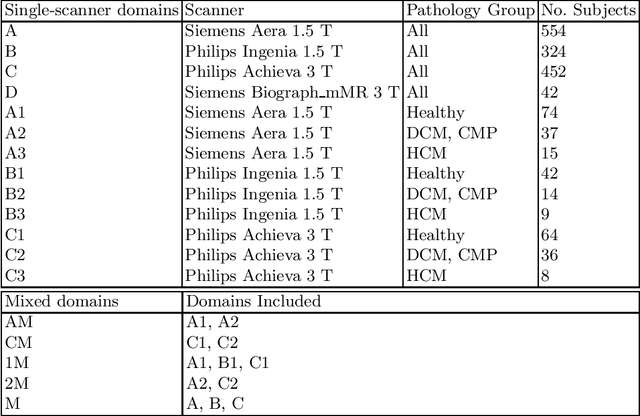
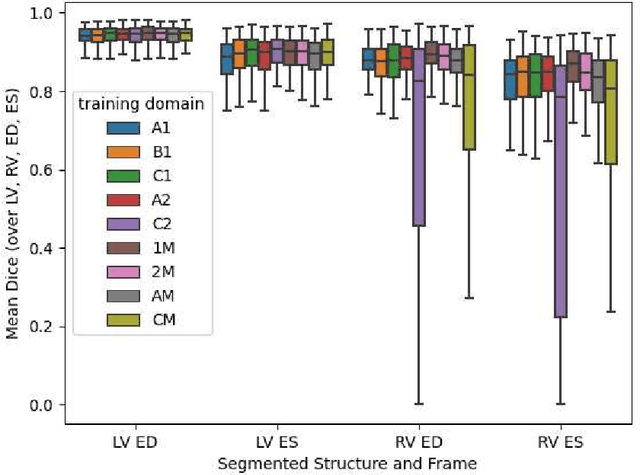
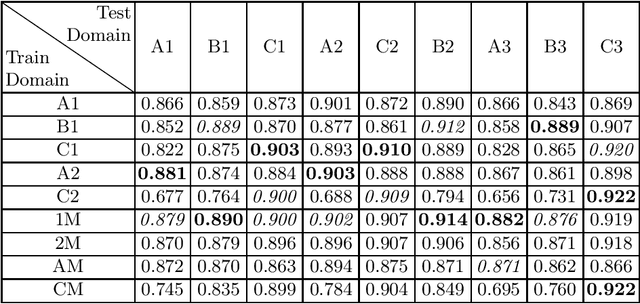
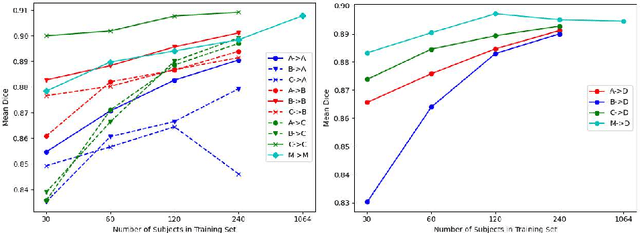
Abstract:Domain shift refers to the difference in the data distribution of two datasets, normally between the training set and the test set for machine learning algorithms. Domain shift is a serious problem for generalization of machine learning models and it is well-established that a domain shift between the training and test sets may cause a drastic drop in the model's performance. In medical imaging, there can be many sources of domain shift such as different scanners or scan protocols, different pathologies in the patient population, anatomical differences in the patient population (e.g. men vs women) etc. Therefore, in order to train models that have good generalization performance, it is important to be aware of the domain shift problem, its potential causes and to devise ways to address it. In this paper, we study the effect of domain shift on left and right ventricle blood pool segmentation in short axis cardiac MR images. Our dataset contains short axis images from 4 different MR scanners and 3 different pathology groups. The training is performed with nnUNet. The results show that scanner differences cause a greater drop in performance compared to changing the pathology group, and that the impact of domain shift is greater on right ventricle segmentation compared to left ventricle segmentation. Increasing the number of training subjects increased cross-scanner performance more than in-scanner performance at small training set sizes, but this difference in improvement decreased with larger training set sizes. Training models using data from multiple scanners improved cross-domain performance.
Uncertainty-Aware Training for Cardiac Resynchronisation Therapy Response Prediction
Sep 22, 2021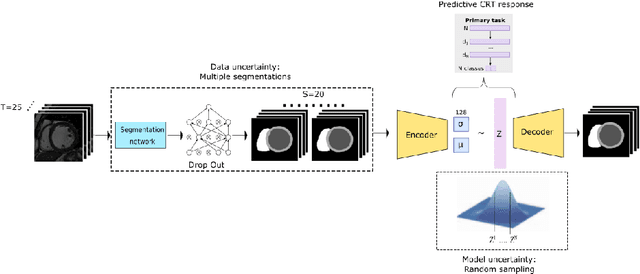
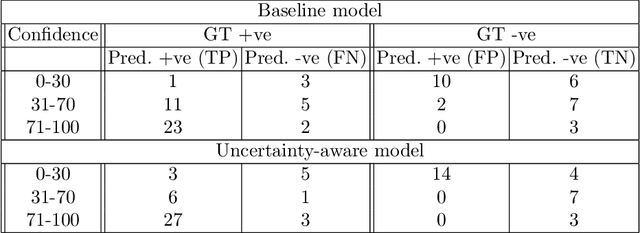
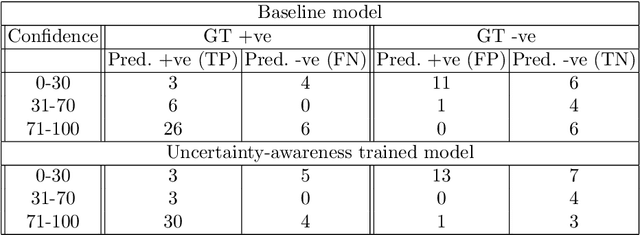
Abstract:Evaluation of predictive deep learning (DL) models beyond conventional performance metrics has become increasingly important for applications in sensitive environments like healthcare. Such models might have the capability to encode and analyse large sets of data but they often lack comprehensive interpretability methods, preventing clinical trust in predictive outcomes. Quantifying uncertainty of a prediction is one way to provide such interpretability and promote trust. However, relatively little attention has been paid to how to include such requirements into the training of the model. In this paper we: (i) quantify the data (aleatoric) and model (epistemic) uncertainty of a DL model for Cardiac Resynchronisation Therapy response prediction from cardiac magnetic resonance images, and (ii) propose and perform a preliminary investigation of an uncertainty-aware loss function that can be used to retrain an existing DL image-based classification model to encourage confidence in correct predictions and reduce confidence in incorrect predictions. Our initial results are promising, showing a significant increase in the (epistemic) confidence of true positive predictions, with some evidence of a reduction in false negative confidence.
Improved AI-based segmentation of apical and basal slices from clinical cine CMR
Sep 20, 2021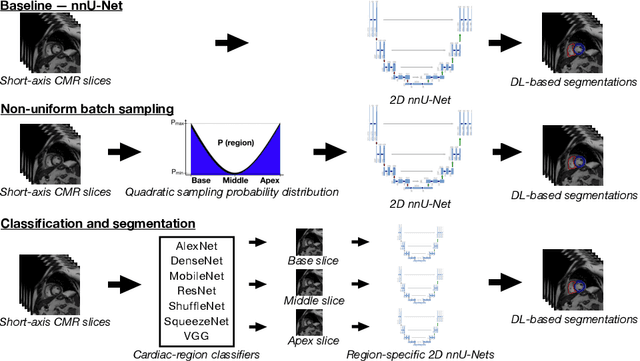
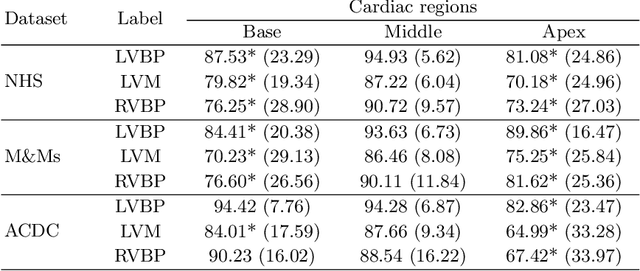
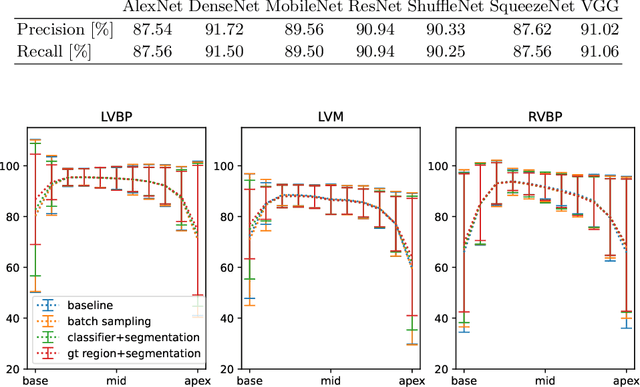
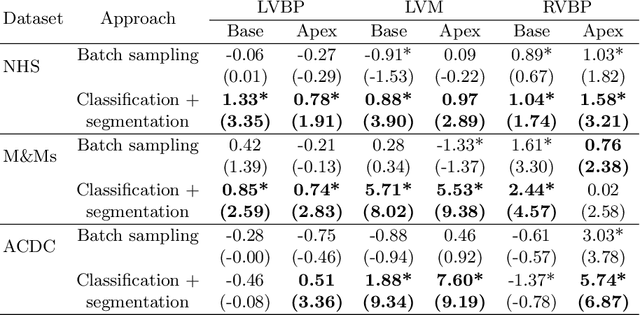
Abstract:Current artificial intelligence (AI) algorithms for short-axis cardiac magnetic resonance (CMR) segmentation achieve human performance for slices situated in the middle of the heart. However, an often-overlooked fact is that segmentation of the basal and apical slices is more difficult. During manual analysis, differences in the basal segmentations have been reported as one of the major sources of disagreement in human interobserver variability. In this work, we aim to investigate the performance of AI algorithms in segmenting basal and apical slices and design strategies to improve their segmentation. We trained all our models on a large dataset of clinical CMR studies obtained from two NHS hospitals (n=4,228) and evaluated them against two external datasets: ACDC (n=100) and M&Ms (n=321). Using manual segmentations as a reference, CMR slices were assigned to one of four regions: non-cardiac, base, middle, and apex. Using the nnU-Net framework as a baseline, we investigated two different approaches to reduce the segmentation performance gap between cardiac regions: (1) non-uniform batch sampling, which allows us to choose how often images from different regions are seen during training; and (2) a cardiac-region classification model followed by three (i.e. base, middle, and apex) region-specific segmentation models. We show that the classification and segmentation approach was best at reducing the performance gap across all datasets. We also show that improvements in the classification performance can subsequently lead to a significantly better performance in the segmentation task.
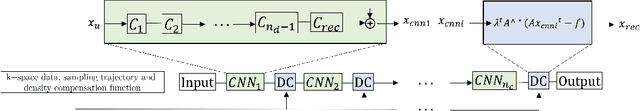
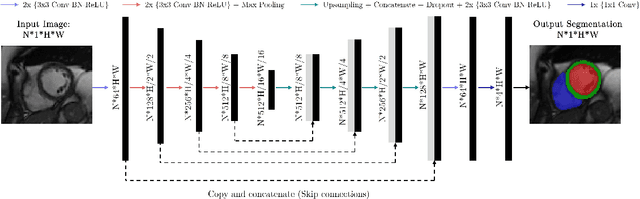
Quality-aware Cine Cardiac MRI Reconstruction and Analysis from Undersampled k-space Data
Sep 16, 2021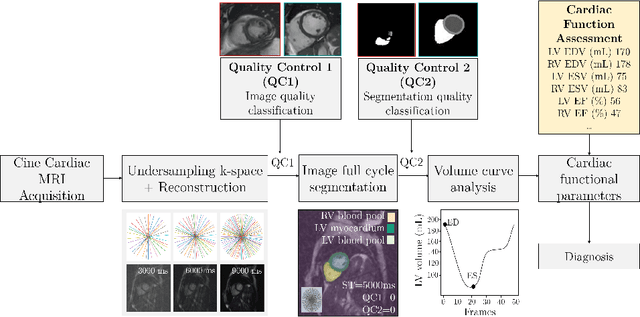
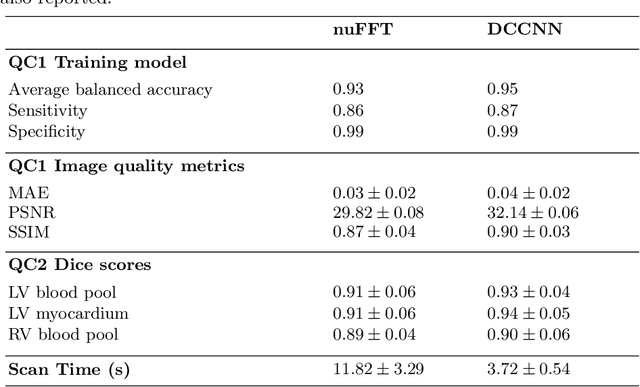
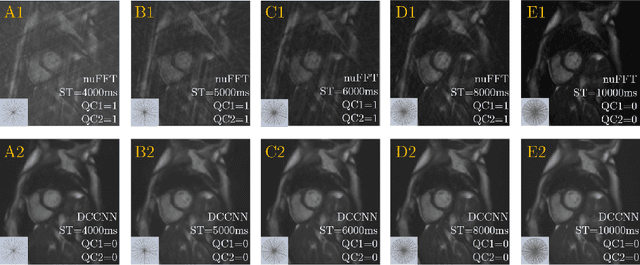
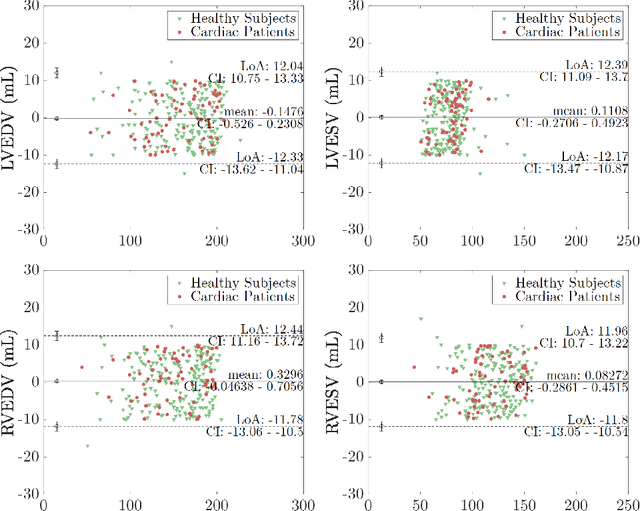
Abstract:Cine cardiac MRI is routinely acquired for the assessment of cardiac health, but the imaging process is slow and typically requires several breath-holds to acquire sufficient k-space profiles to ensure good image quality. Several undersampling-based reconstruction techniques have been proposed during the last decades to speed up cine cardiac MRI acquisition. However, the undersampling factor is commonly fixed to conservative values before acquisition to ensure diagnostic image quality, potentially leading to unnecessarily long scan times. In this paper, we propose an end-to-end quality-aware cine short-axis cardiac MRI framework that combines image acquisition and reconstruction with downstream tasks such as segmentation, volume curve analysis and estimation of cardiac functional parameters. The goal is to reduce scan time by acquiring only a fraction of k-space data to enable the reconstruction of images that can pass quality control checks and produce reliable estimates of cardiac functional parameters. The framework consists of a deep learning model for the reconstruction of 2D+t cardiac cine MRI images from undersampled data, an image quality-control step to detect good quality reconstructions, followed by a deep learning model for bi-ventricular segmentation, a quality-control step to detect good quality segmentations and automated calculation of cardiac functional parameters. To demonstrate the feasibility of the proposed approach, we perform simulations using a cohort of selected participants from the UK Biobank (n=270), 200 healthy subjects and 70 patients with cardiomyopathies. Our results show that we can produce quality-controlled images in a scan time reduced from 12 to 4 seconds per slice, enabling reliable estimates of cardiac functional parameters such as ejection fraction within 5% mean absolute error.
A Multimodal Deep Learning Model for Cardiac Resynchronisation Therapy Response Prediction
Jul 20, 2021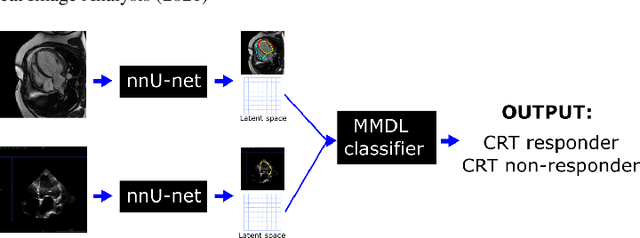
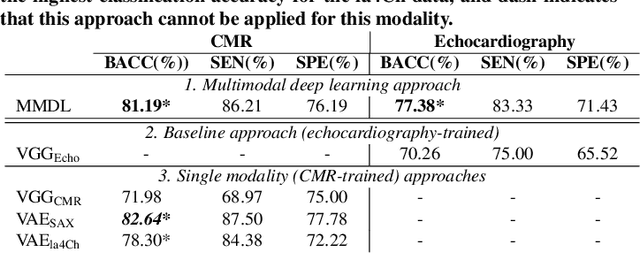
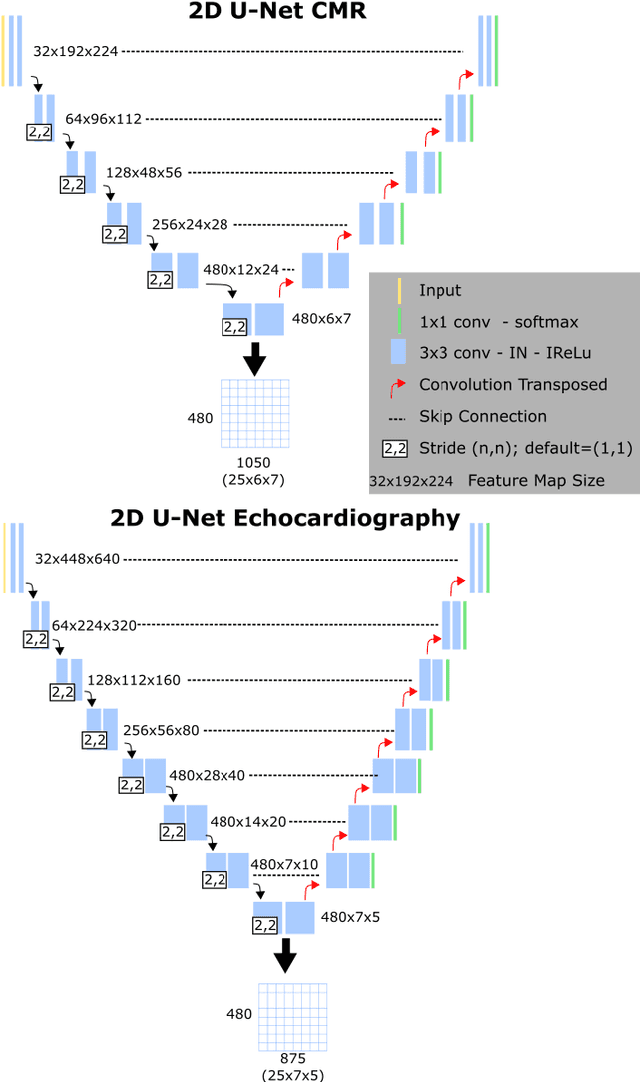
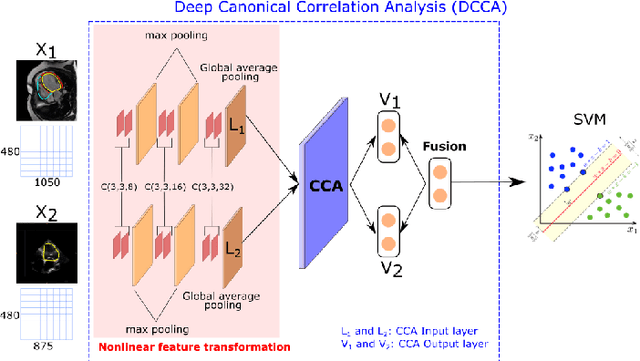
Abstract:We present a novel multimodal deep learning framework for cardiac resynchronisation therapy (CRT) response prediction from 2D echocardiography and cardiac magnetic resonance (CMR) data. The proposed method first uses the `nnU-Net' segmentation model to extract segmentations of the heart over the full cardiac cycle from the two modalities. Next, a multimodal deep learning classifier is used for CRT response prediction, which combines the latent spaces of the segmentation models of the two modalities. At inference time, this framework can be used with 2D echocardiography data only, whilst taking advantage of the implicit relationship between CMR and echocardiography features learnt from the model. We evaluate our pipeline on a cohort of 50 CRT patients for whom paired echocardiography/CMR data were available, and results show that the proposed multimodal classifier results in a statistically significant improvement in accuracy compared to the baseline approach that uses only 2D echocardiography data. The combination of multimodal data enables CRT response to be predicted with 77.38% accuracy (83.33% sensitivity and 71.43% specificity), which is comparable with the current state-of-the-art in machine learning-based CRT response prediction. Our work represents the first multimodal deep learning approach for CRT response prediction.
Fairness in Cardiac MR Image Analysis: An Investigation of Bias Due to Data Imbalance in Deep Learning Based Segmentation
Jul 01, 2021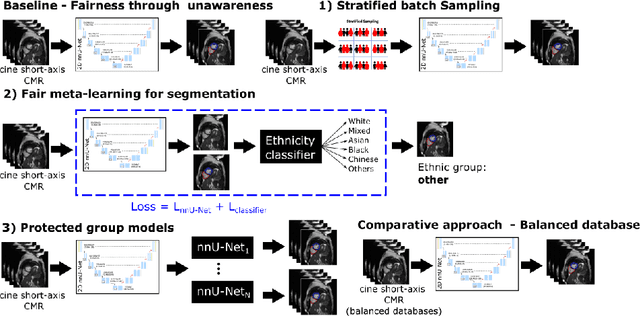
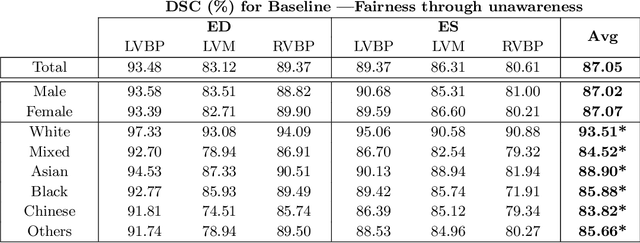
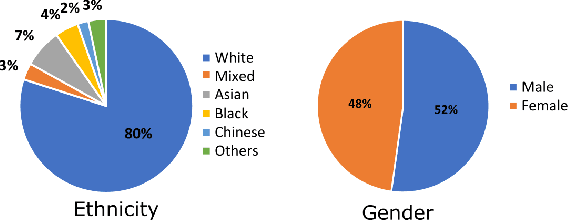
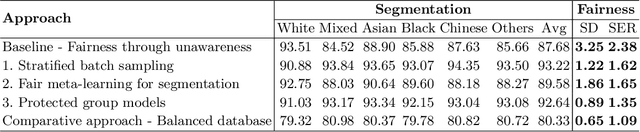
Abstract:The subject of "fairness" in artificial intelligence (AI) refers to assessing AI algorithms for potential bias based on demographic characteristics such as race and gender, and the development of algorithms to address this bias. Most applications to date have been in computer vision, although some work in healthcare has started to emerge. The use of deep learning (DL) in cardiac MR segmentation has led to impressive results in recent years, and such techniques are starting to be translated into clinical practice. However, no work has yet investigated the fairness of such models. In this work, we perform such an analysis for racial/gender groups, focusing on the problem of training data imbalance, using a nnU-Net model trained and evaluated on cine short axis cardiac MR data from the UK Biobank dataset, consisting of 5,903 subjects from 6 different racial groups. We find statistically significant differences in Dice performance between different racial groups. To reduce the racial bias, we investigated three strategies: (1) stratified batch sampling, in which batch sampling is stratified to ensure balance between racial groups; (2) fair meta-learning for segmentation, in which a DL classifier is trained to classify race and jointly optimized with the segmentation model; and (3) protected group models, in which a different segmentation model is trained for each racial group. We also compared the results to the scenario where we have a perfectly balanced database. To assess fairness we used the standard deviation (SD) and skewed error ratio (SER) of the average Dice values. Our results demonstrate that the racial bias results from the use of imbalanced training data, and that all proposed bias mitigation strategies improved fairness, with the best SD and SER resulting from the use of protected group models.
Channel Attention Networks for Robust MR Fingerprinting Matching
Dec 02, 2020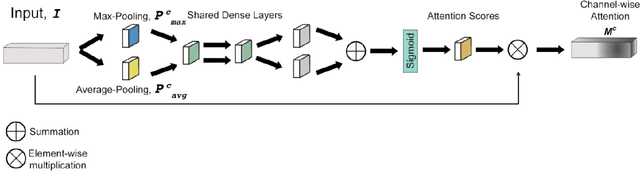
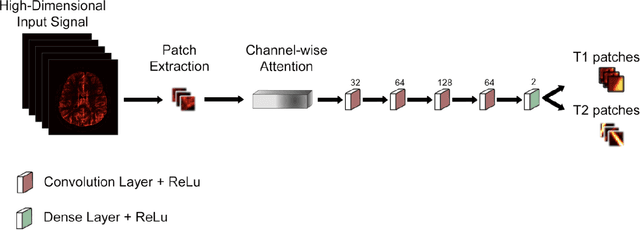
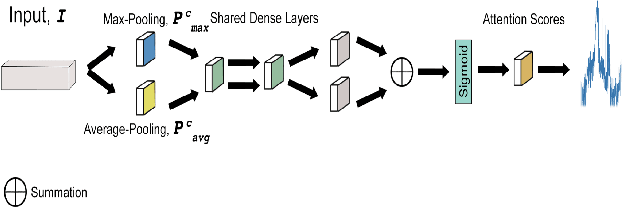
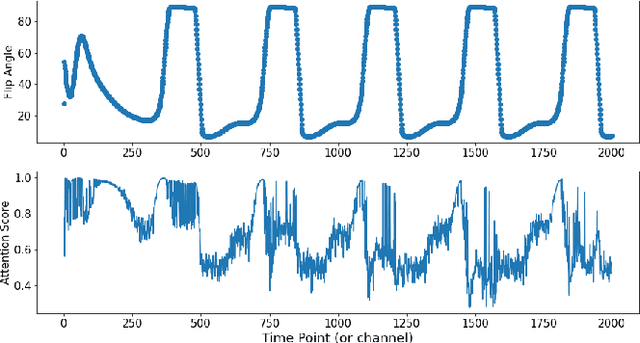
Abstract:Magnetic Resonance Fingerprinting (MRF) enables simultaneous mapping of multiple tissue parameters such as T1 and T2 relaxation times. The working principle of MRF relies on varying acquisition parameters pseudo-randomly, so that each tissue generates its unique signal evolution during scanning. Even though MRF provides faster scanning, it has disadvantages such as erroneous and slow generation of the corresponding parametric maps, which needs to be improved. Moreover, there is a need for explainable architectures for understanding the guiding signals to generate accurate parametric maps. In this paper, we addressed both of these shortcomings by proposing a novel neural network architecture consisting of a channel-wise attention module and a fully convolutional network. The proposed approach, evaluated over 3 simulated MRF signals, reduces error in the reconstruction of tissue parameters by 8.88% for T1 and 75.44% for T2 with respect to state-of-the-art methods. Another contribution of this study is a new channel selection method: attention-based channel selection. Furthermore, the effect of patch size and temporal frames of MRF signal on channel reduction are analyzed by employing a channel-wise attention.
Deep Learning for Automatic Spleen Length Measurement in Sickle Cell Disease Patients
Sep 06, 2020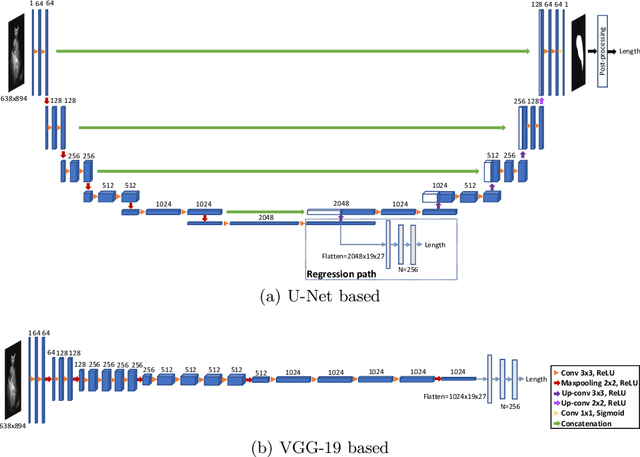
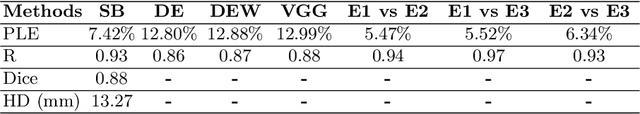
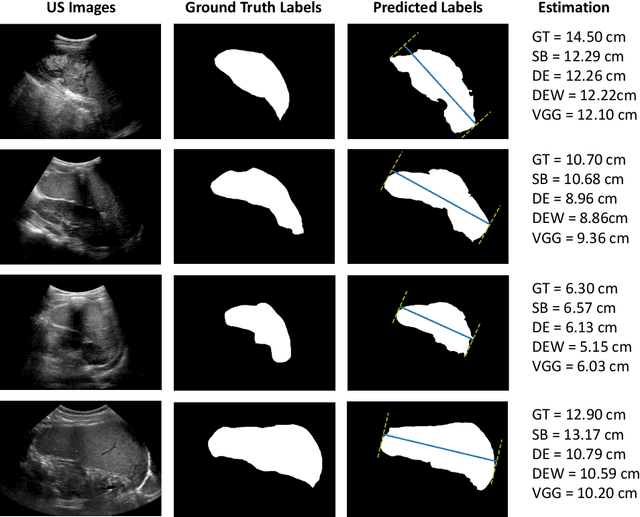
Abstract:Sickle Cell Disease (SCD) is one of the most common genetic diseases in the world. Splenomegaly (abnormal enlargement of the spleen) is frequent among children with SCD. If left untreated, splenomegaly can be life-threatening. The current workflow to measure spleen size includes palpation, possibly followed by manual length measurement in 2D ultrasound imaging. However, this manual measurement is dependent on operator expertise and is subject to intra- and inter-observer variability. We investigate the use of deep learning to perform automatic estimation of spleen length from ultrasound images. We investigate two types of approach, one segmentation-based and one based on direct length estimation, and compare the results against measurements made by human experts. Our best model (segmentation-based) achieved a percentage length error of 7.42%, which is approaching the level of inter-observer variability (5.47%-6.34%). To the best of our knowledge, this is the first attempt to measure spleen size in a fully automated way from ultrasound images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge