Amin Katouzian
Pneumothorax and chest tube classification on chest x-rays for detection of missed pneumothorax
Nov 14, 2020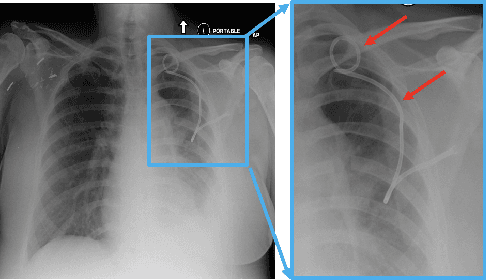
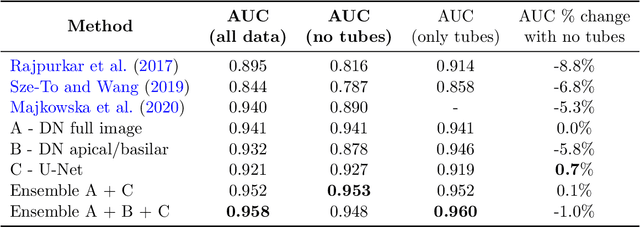
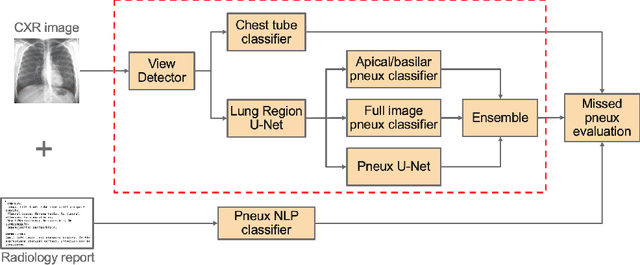
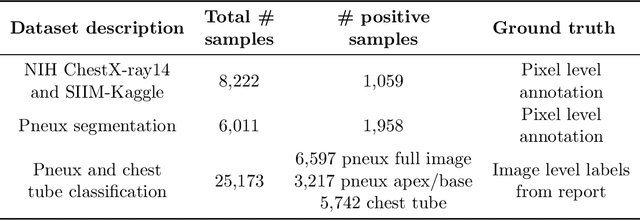
Abstract:Chest x-ray imaging is widely used for the diagnosis of pneumothorax and there has been significant interest in developing automated methods to assist in image interpretation. We present an image classification pipeline which detects pneumothorax as well as the various types of chest tubes that are commonly used to treat pneumothorax. Our multi-stage algorithm is based on lung segmentation followed by pneumothorax classification, including classification of patches that are most likely to contain pneumothorax. This algorithm achieves state of the art performance for pneumothorax classification on an open-source benchmark dataset. Unlike previous work, this algorithm shows comparable performance on data with and without chest tubes and thus has an improved clinical utility. To evaluate these algorithms in a realistic clinical scenario, we demonstrate the ability to identify real cases of missed pneumothorax in a large dataset of chest x-ray studies.
ReLayNet: Retinal Layer and Fluid Segmentation of Macular Optical Coherence Tomography using Fully Convolutional Network
Jul 07, 2017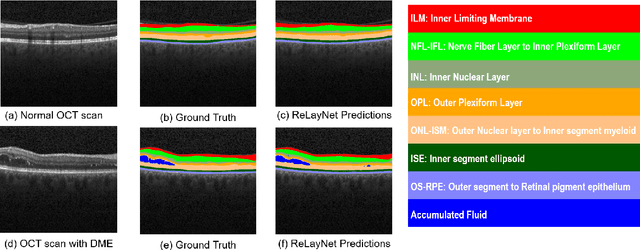
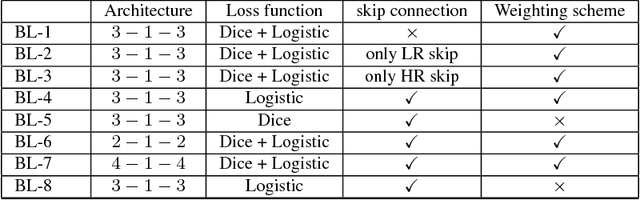
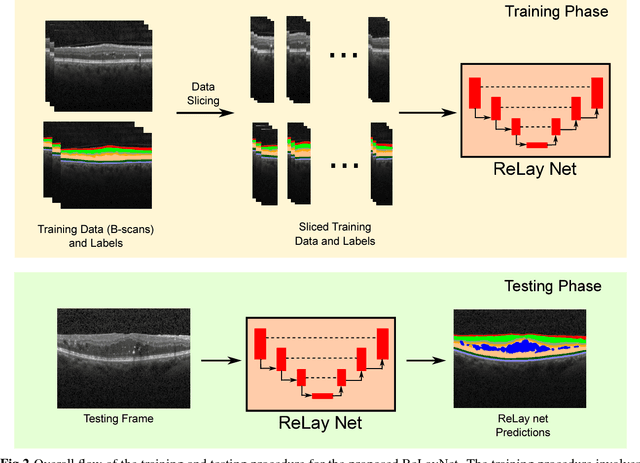
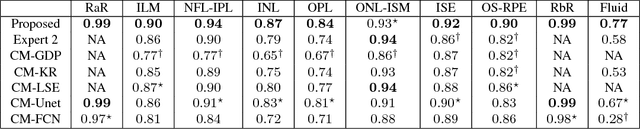
Abstract:Optical coherence tomography (OCT) is used for non-invasive diagnosis of diabetic macular edema assessing the retinal layers. In this paper, we propose a new fully convolutional deep architecture, termed ReLayNet, for end-to-end segmentation of retinal layers and fluid masses in eye OCT scans. ReLayNet uses a contracting path of convolutional blocks (encoders) to learn a hierarchy of contextual features, followed by an expansive path of convolutional blocks (decoders) for semantic segmentation. ReLayNet is trained to optimize a joint loss function comprising of weighted logistic regression and Dice overlap loss. The framework is validated on a publicly available benchmark dataset with comparisons against five state-of-the-art segmentation methods including two deep learning based approaches to substantiate its effectiveness.
Error Corrective Boosting for Learning Fully Convolutional Networks with Limited Data
Jul 02, 2017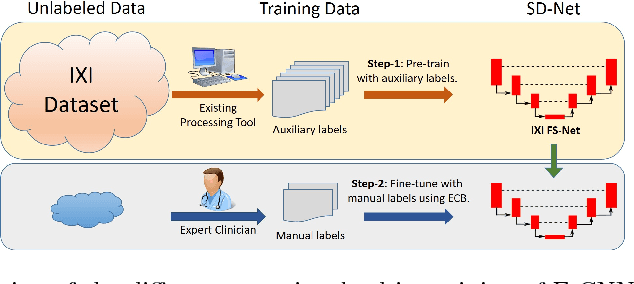
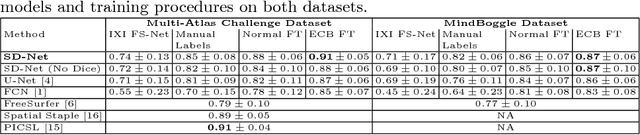
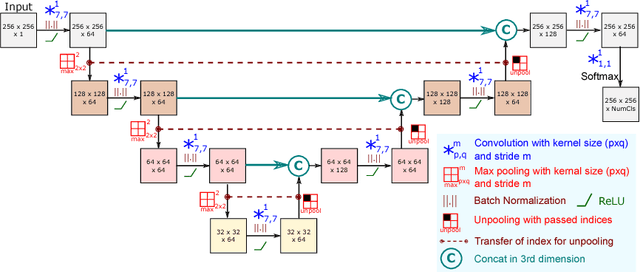
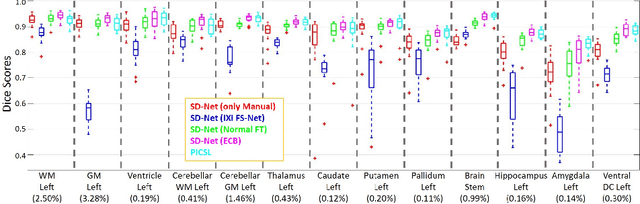
Abstract:Training deep fully convolutional neural networks (F-CNNs) for semantic image segmentation requires access to abundant labeled data. While large datasets of unlabeled image data are available in medical applications, access to manually labeled data is very limited. We propose to automatically create auxiliary labels on initially unlabeled data with existing tools and to use them for pre-training. For the subsequent fine-tuning of the network with manually labeled data, we introduce error corrective boosting (ECB), which emphasizes parameter updates on classes with lower accuracy. Furthermore, we introduce SkipDeconv-Net (SD-Net), a new F-CNN architecture for brain segmentation that combines skip connections with the unpooling strategy for upsampling. The SD-Net addresses challenges of severe class imbalance and errors along boundaries. With application to whole-brain MRI T1 scan segmentation, we generate auxiliary labels on a large dataset with FreeSurfer and fine-tune on two datasets with manual annotations. Our results show that the inclusion of auxiliary labels and ECB yields significant improvements. SD-Net segments a 3D scan in 7 secs in comparison to 30 hours for the closest multi-atlas segmentation method, while reaching similar performance. It also outperforms the latest state-of-the-art F-CNN models.
Learning Robust Hash Codes for Multiple Instance Image Retrieval
Mar 16, 2017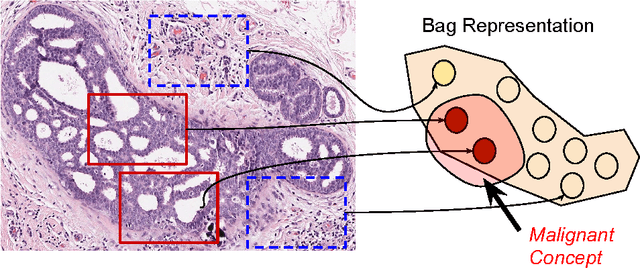
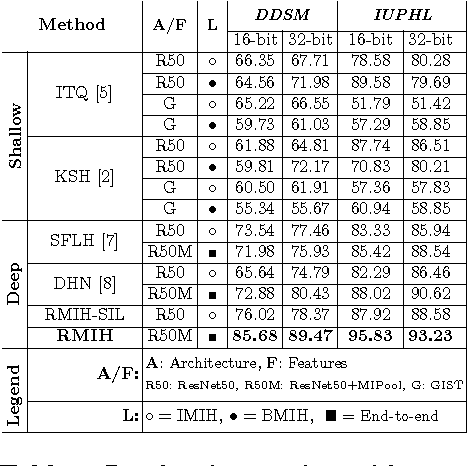
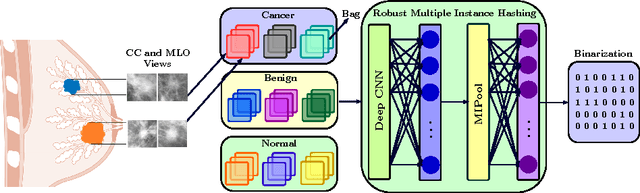
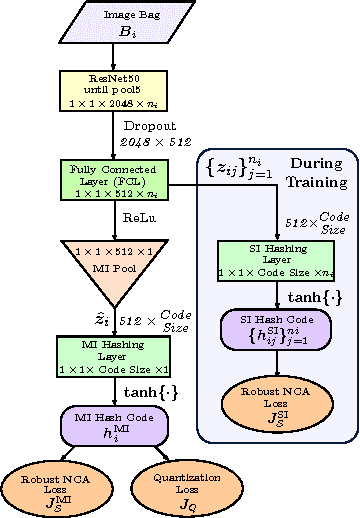
Abstract:In this paper, for the first time, we introduce a multiple instance (MI) deep hashing technique for learning discriminative hash codes with weak bag-level supervision suited for large-scale retrieval. We learn such hash codes by aggregating deeply learnt hierarchical representations across bag members through a dedicated MI pool layer. For better trainability and retrieval quality, we propose a two-pronged approach that includes robust optimization and training with an auxiliary single instance hashing arm which is down-regulated gradually. We pose retrieval for tumor assessment as an MI problem because tumors often coexist with benign masses and could exhibit complementary signatures when scanned from different anatomical views. Experimental validations on benchmark mammography and histology datasets demonstrate improved retrieval performance over the state-of-the-art methods.
Cross-Modal Manifold Learning for Cross-modal Retrieval
Dec 19, 2016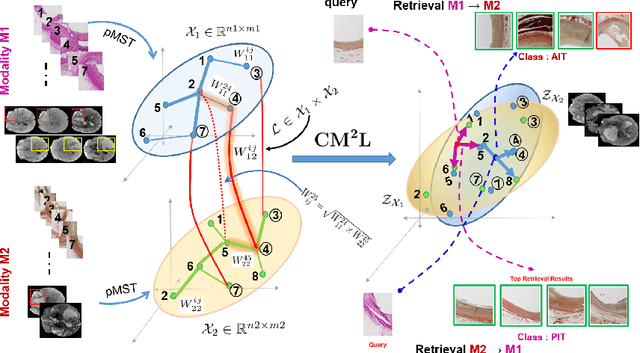
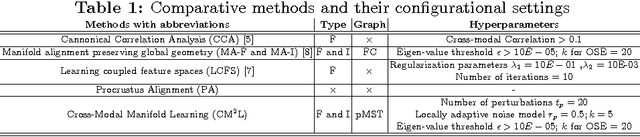
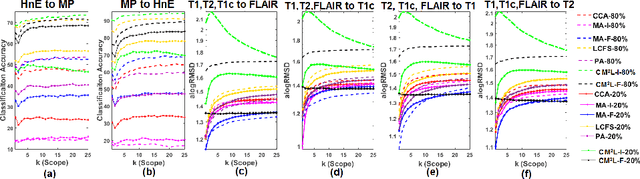
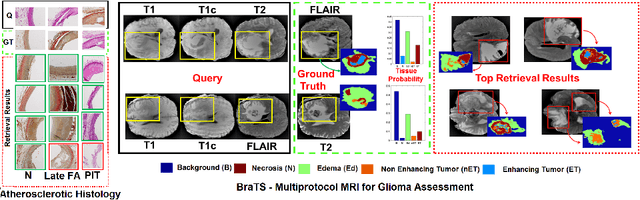
Abstract:This paper presents a new scalable algorithm for cross-modal similarity preserving retrieval in a learnt manifold space. Unlike existing approaches that compromise between preserving global and local geometries, the proposed technique respects both simultaneously during manifold alignment. The global topologies are maintained by recovering underlying mapping functions in the joint manifold space by deploying partially corresponding instances. The inter-, and intra-modality affinity matrices are then computed to reinforce original data skeleton using perturbed minimum spanning tree (pMST), and maximizing the affinity among similar cross-modal instances, respectively. The performance of proposed algorithm is evaluated upon two multimodal image datasets (coronary atherosclerosis histology and brain MRI) for two applications: classification, and regression. Our exhaustive validations and results demonstrate the superiority of our technique over comparative methods and its feasibility for improving computer-assisted diagnosis systems, where disease-specific complementary information shall be aggregated and interpreted across modalities to form the final decision.
Deep Residual Hashing
Dec 16, 2016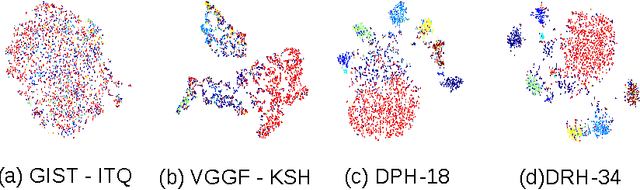
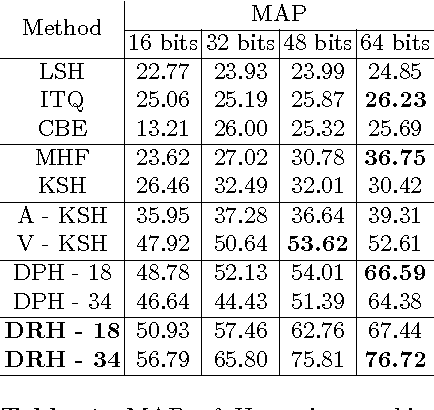
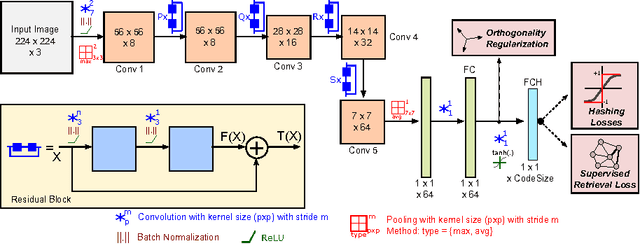
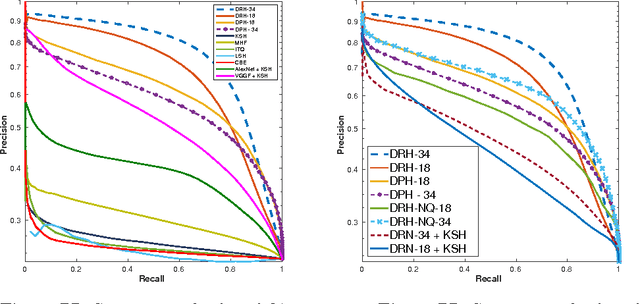
Abstract:Hashing aims at generating highly compact similarity preserving code words which are well suited for large-scale image retrieval tasks. Most existing hashing methods first encode the images as a vector of hand-crafted features followed by a separate binarization step to generate hash codes. This two-stage process may produce sub-optimal encoding. In this paper, for the first time, we propose a deep architecture for supervised hashing through residual learning, termed Deep Residual Hashing (DRH), for an end-to-end simultaneous representation learning and hash coding. The DRH model constitutes four key elements: (1) a sub-network with multiple stacked residual blocks; (2) hashing layer for binarization; (3) supervised retrieval loss function based on neighbourhood component analysis for similarity preserving embedding; and (4) hashing related losses and regularisation to control the quantization error and improve the quality of hash coding. We present results of extensive experiments on a large public chest x-ray image database with co-morbidities and discuss the outcome showing substantial improvements over the latest state-of-the art methods.
An Efficient Training Algorithm for Kernel Survival Support Vector Machines
Nov 21, 2016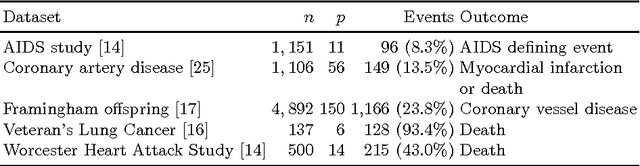
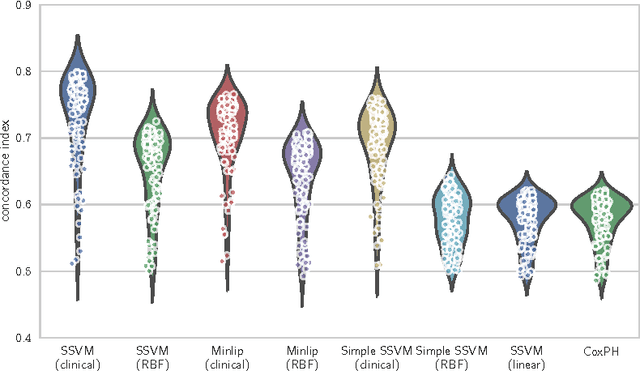
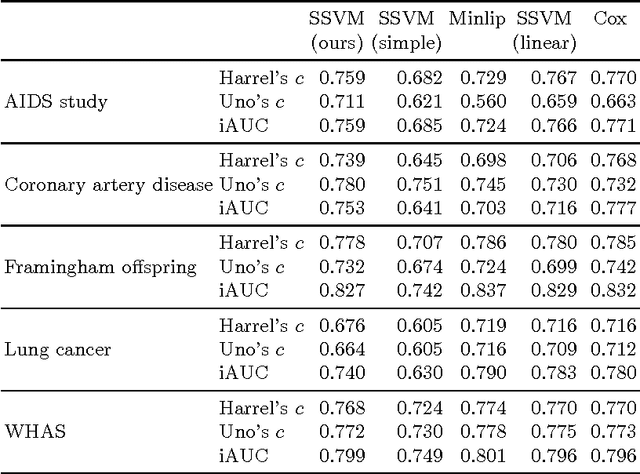
Abstract:Survival analysis is a fundamental tool in medical research to identify predictors of adverse events and develop systems for clinical decision support. In order to leverage large amounts of patient data, efficient optimisation routines are paramount. We propose an efficient training algorithm for the kernel survival support vector machine (SSVM). We directly optimise the primal objective function and employ truncated Newton optimisation and order statistic trees to significantly lower computational costs compared to previous training algorithms, which require $O(n^4)$ space and $O(p n^6)$ time for datasets with $n$ samples and $p$ features. Our results demonstrate that our proposed optimisation scheme allows analysing data of a much larger scale with no loss in prediction performance. Experiments on synthetic and 5 real-world datasets show that our technique outperforms existing kernel SSVM formulations if the amount of right censoring is high ($\geq85\%$), and performs comparably otherwise.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge