Ali Erturk
SELMA3D challenge: Self-supervised learning for 3D light-sheet microscopy image segmentation
Jan 07, 2025Abstract:Recent innovations in light sheet microscopy, paired with developments in tissue clearing techniques, enable the 3D imaging of large mammalian tissues with cellular resolution. Combined with the progress in large-scale data analysis, driven by deep learning, these innovations empower researchers to rapidly investigate the morphological and functional properties of diverse biological samples. Segmentation, a crucial preliminary step in the analysis process, can be automated using domain-specific deep learning models with expert-level performance. However, these models exhibit high sensitivity to domain shifts, leading to a significant drop in accuracy when applied to data outside their training distribution. To address this limitation, and inspired by the recent success of self-supervised learning in training generalizable models, we organized the SELMA3D Challenge during the MICCAI 2024 conference. SELMA3D provides a vast collection of light-sheet images from cleared mice and human brains, comprising 35 large 3D images-each with over 1000^3 voxels-and 315 annotated small patches for finetuning, preliminary testing and final testing. The dataset encompasses diverse biological structures, including vessel-like and spot-like structures. Five teams participated in all phases of the challenge, and their proposed methods are reviewed in this paper. Quantitative and qualitative results from most participating teams demonstrate that self-supervised learning on large datasets improves segmentation model performance and generalization. We will continue to support and extend SELMA3D as an inaugural MICCAI challenge focused on self-supervised learning for 3D microscopy image segmentation.
blob loss: instance imbalance aware loss functions for semantic segmentation
May 17, 2022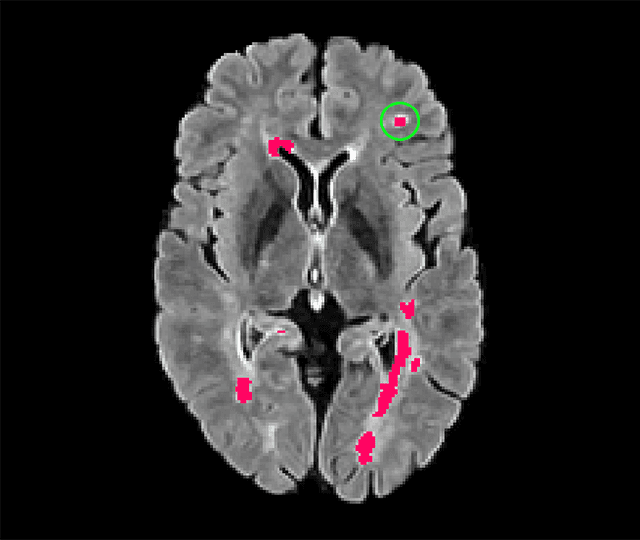
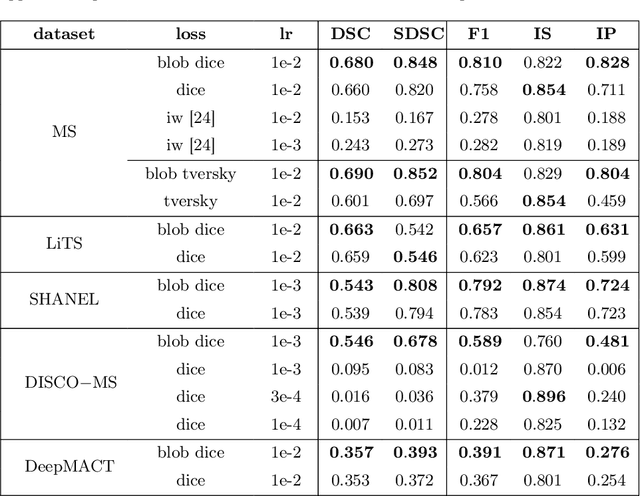

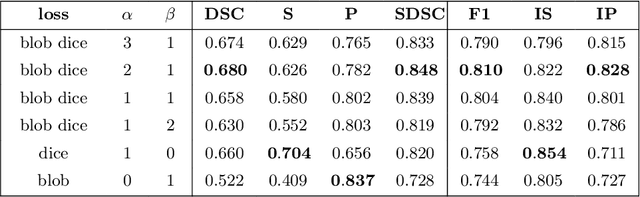
Abstract:Deep convolutional neural networks have proven to be remarkably effective in semantic segmentation tasks. Most popular loss functions were introduced targeting improved volumetric scores, such as the Sorensen Dice coefficient. By design, DSC can tackle class imbalance; however, it does not recognize instance imbalance within a class. As a result, a large foreground instance can dominate minor instances and still produce a satisfactory Sorensen Dice coefficient. Nevertheless, missing out on instances will lead to poor detection performance. This represents a critical issue in applications such as disease progression monitoring. For example, it is imperative to locate and surveil small-scale lesions in the follow-up of multiple sclerosis patients. We propose a novel family of loss functions, nicknamed blob loss, primarily aimed at maximizing instance-level detection metrics, such as F1 score and sensitivity. Blob loss is designed for semantic segmentation problems in which the instances are the connected components within a class. We extensively evaluate a DSC-based blob loss in five complex 3D semantic segmentation tasks featuring pronounced instance heterogeneity in terms of texture and morphology. Compared to soft Dice loss, we achieve 5 percent improvement for MS lesions, 3 percent improvement for liver tumor, and an average 2 percent improvement for Microscopy segmentation tasks considering F1 score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge